Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions
- PMID: 19640992
- PMCID: PMC2748034
- DOI: 10.1128/JVI.00571-09
Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions
Abstract
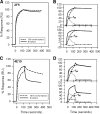
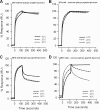
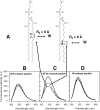
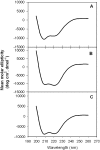
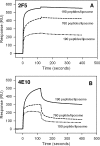
The binding of neutralizing antibodies 2F5 and 4E10 to human immunodeficiency virus type 1 (HIV-1) gp41 involves both the viral membrane and gp41 membrane proximal external region (MPER) epitopes. In this study, we have used several biophysical tools to examine the secondary structure, orientation, and depth of immersion of gp41 MPER peptides in liposomes and to determine how the orientation of the MPER with lipids affects the binding kinetics of monoclonal antibodies (MAbs) 2F5 and 4E10. The binding of 2F5 and 4E10 both to their respective nominal epitopes and to a biepitope (includes 2F5 and 4E10 epitopes) MPER peptide-liposome conjugate was best described by a two-step encounter-docking model. Analysis of the binding kinetics and the effect of temperature on the binding stability of 2F5 and 4E10 to MPER peptide-liposome conjugates revealed that the docking of 4E10 was relatively slower and thermodynamically less favorable. The results of fluorescence-quenching and fluorescence resonance energy transfer experiments showed that the 2F5 epitope was more solvent exposed, whereas the 4E10 epitope was immersed in the polar-apolar interfacial region of the lipid bilayer. A circular dichroism spectroscopic study demonstrated that the nominal epitope and biepitope MPER peptides adopted ordered structures with differing helical contents when anchored to liposomes. Furthermore, anchoring of MPER peptides to the membrane via a hydrophobic anchor sequence was required for efficient MAb docking. These results support the model that the ability of 2F5 and 4E10 to bind to membrane lipid is required for stable docking to membrane-embedded MPER residues. These data have important implications for the design and use of peptide-liposome conjugates as immunogens for the induction of MPER-neutralizing antibodies.
Figures

References
-
- Alam, S. M., S. Dennison, M. Morelli, H. Liao, R. Zhang, B. F. Haynes, S. C. Harrison, and B. Chen. 2008. The role of lipid reactivity of HIV-1 gp41 membrane proximal antibodies in neutralization of HIV-1: novel liposome based immunogen design, abstr. OA07 to 06. Abstracts from AIDS Vaccine 2008, Cape Town, South Africa. October 13-16, 2008. AIDS Res. Hum Retrovir. 24(Suppl. 1):23-24.
-
- Alam, S. M., M. McAdams, D. Boren, M. Rak, R. M. Scearce, F. Gao, Z. T. Camacho, D. Gewirth, G. Kelsoe, P. Chen, and B. F. Haynes. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424-4435. - PMC - PubMed
-
- Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. - PubMed
-
- Barenholz, Y., D. Gibbes, B. J. Litman, J. Goll, T. E. Thompson, and R. D. Carlson. 1977. A simple method for the preparation of homogeneous phospholipid vesicles. Biochemistry 16:2806-2810. - PubMed
-
- Bolen, E. J., and P. W. Holloway. 1990. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry 29:9638-9643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

