Mapping of functional domains in herpesvirus saimiri complement control protein homolog: complement control protein domain 2 is the smallest structural unit displaying cofactor and decay-accelerating activities
- PMID: 19640995
- PMCID: PMC2748004
- DOI: 10.1128/JVI.00217-09
Mapping of functional domains in herpesvirus saimiri complement control protein homolog: complement control protein domain 2 is the smallest structural unit displaying cofactor and decay-accelerating activities
Abstract
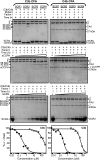
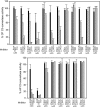
Herpesvirus saimiri encodes a functional homolog of human regulator-of-complement-activation proteins named CCPH that inactivates complement by accelerating the decay of C3 convertases and by serving as a cofactor in factor I-mediated inactivation of their subunits C3b and C4b. Here, we map the functional domains of CCPH. We demonstrate that short consensus repeat 2 (SCR2) is the minimum domain essential for classical/lectin pathway C3 convertase decay-accelerating activity as well as for factor I cofactor activity for C3b and C4b. Thus, CCPH is the first example wherein a single SCR domain has been shown to display complement regulatory functions.
Figures



Similar articles
-
Dissection of functional sites in herpesvirus saimiri complement control protein homolog.J Virol. 2013 Jan;87(1):282-95. doi: 10.1128/JVI.01867-12. Epub 2012 Oct 17. J Virol. 2013. PMID: 23077301 Free PMC article.
-
Functional characterization of the complement control protein homolog of herpesvirus saimiri: ARG-118 is critical for factor I cofactor activities.J Biol Chem. 2006 Aug 11;281(32):23119-28. doi: 10.1074/jbc.M603085200. Epub 2006 Jun 7. J Biol Chem. 2006. PMID: 16760474
-
Identification of complement regulatory domains in vaccinia virus complement control protein.J Virol. 2005 Oct;79(19):12382-93. doi: 10.1128/JVI.79.19.12382-12393.2005. J Virol. 2005. PMID: 16160165 Free PMC article.
-
Structure-function relationships of complement receptor type 1.Immunol Rev. 2001 Apr;180:112-22. doi: 10.1034/j.1600-065x.2001.1800110.x. Immunol Rev. 2001. PMID: 11414353 Review.
-
Structural insights on complement activation.FEBS J. 2015 Oct;282(20):3883-91. doi: 10.1111/febs.13399. Epub 2015 Aug 31. FEBS J. 2015. PMID: 26250513 Review.
Cited by
-
Complement Evasion Strategies of Viruses: An Overview.Front Microbiol. 2017 Jun 16;8:1117. doi: 10.3389/fmicb.2017.01117. eCollection 2017. Front Microbiol. 2017. PMID: 28670306 Free PMC article. Review.
-
Virus-Encoded Complement Regulators: Current Status.Viruses. 2021 Jan 29;13(2):208. doi: 10.3390/v13020208. Viruses. 2021. PMID: 33573085 Free PMC article. Review.
-
Dissection of functional sites in herpesvirus saimiri complement control protein homolog.J Virol. 2013 Jan;87(1):282-95. doi: 10.1128/JVI.01867-12. Epub 2012 Oct 17. J Virol. 2013. PMID: 23077301 Free PMC article.
-
Viral-derived complement inhibitors: current status and potential role in immunomodulation.Exp Biol Med (Maywood). 2017 Feb;242(4):397-410. doi: 10.1177/1535370216675772. Epub 2016 Oct 26. Exp Biol Med (Maywood). 2017. PMID: 27798122 Free PMC article. Review.
References
-
- Ahmad, M., K. Pyaram, J. Mullick, and A. Sahu. 2007. Viral complement regulators: the expert mimicking swindlers. Indian J. Biochem. Biophys. 44:331-343. - PubMed
-
- Blom, A. M., L. Kask, and B. Dahlback. 2001. Structural requirements for the complement regulatory activities of C4BP. J. Biol. Chem. 276:27136-27144. - PubMed
-
- Bramley, J. C., A. Davies, and P. J. Lachmann. 1997. Herpesvirus saimiri CD59—baculovirus expression and characterisation of complement inhibitory activity. Biochem. Soc. Trans. 25:354S. - PubMed
-
- Brodbeck, W. G., D. Liu, J. Sperry, C. Mold, and M. E. Medof. 1996. Localization of classical and alternative pathway regulatory activity within the decay-accelerating factor. J. Immunol. 156:2528-2533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

