Novel epigenetic mechanisms that control pluripotency and quiescence of adult bone marrow-derived Oct4(+) very small embryonic-like stem cells
- PMID: 19641521
- PMCID: PMC2783188
- DOI: 10.1038/leu.2009.153
Novel epigenetic mechanisms that control pluripotency and quiescence of adult bone marrow-derived Oct4(+) very small embryonic-like stem cells
Abstract
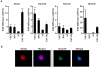
Recently, we identified in adult tissues a population of Oct4(+)SSEA-1(+)Sca-1(+)lin(-)CD45(-) very small embryonic-like stem cells (VSELs). First, to address recent controversies on Oct4 expression in cells isolated from adult organs, we show here evidence that Oct4 promoter in bone marrow (BM)-derived VSELs has an open chromatin structure and is actively transcribed. Next, to explain VSELs quiescence and lack of teratoma formation, we demonstrate a unique DNA methylation pattern at some developmentally crucial imprinted genes, showing hypomethylation/erasure of imprints in paternally methylated and hypermethylation of imprints in maternally methylated ones. These epigenetic characteristics leading to upregulation in VSELs of H19 and p57(KIP2) (also known as Cdkn1c) and repression of Igf2 and Rasgrf1 explain VSEL's quiescent status. Interestingly, this unique pattern in imprinted gene methylation is reverted in cocultures with a C2C12 supportive cell-line when VSELs are induced to form VSEL-derived spheres (VSEL-DSs) enriched for stem cells able to differentiate into all three germ layers. Therefore, we suggest that the proliferative/developmental potential of Oct4(+) VSELs is epigenetically regulated by expression of Oct4 and some imprinted genes, and postulate that restoring the proper methylation pattern of imprinted genes will be a crucial step for using these cells in regenerative medicine.
Conflict of interest statement
Figures




References
-
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
-
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. - PubMed
-
- Matsui Y, Zsebo K, Hogan BLM. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. - PubMed
-
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. - PubMed
-
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

