The mRNA expression of SATB1 and SATB2 in human breast cancer
- PMID: 19642980
- PMCID: PMC2731048
- DOI: 10.1186/1475-2867-9-18
The mRNA expression of SATB1 and SATB2 in human breast cancer
Abstract
Background: SATB1 is a nuclear protein that has been recently reported to be a 'genome organizer' which delineates specific epigenetic modifications at target gene loci, directly up-regulating metastasis-associated genes while down-regulating tumor-suppressor genes. In this study, the level of mRNA expression of SATB1 and SATB2 were assessed in normal and malignant breast tissue in a cohort of women with breast cancer and correlated to conventional clinico-pathological parameters.
Materials and methods: Breast cancer tissues (n = 115) and normal background tissues (n = 31) were collected immediately after excision during surgery. Following RNA extraction, reverse transcription was carried out and transcript levels were determined using real-time quantitative PCR and normalized against beta-actin expression. Transcript levels within the breast cancer specimens were compared to the normal background tissues and analyzed against TNM stage, nodal involvement, tumour grade and clinical outcome over a 10 year follow-up period.
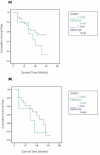
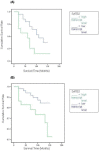
Results: The levels of SATB1 were higher in malignant compared with normal breast tissue (p = 0.0167). SATB1 expression increased with increasing TNM stage (TNM1 vs. TNM2 p = 0.0264), increasing tumour grade (grade1 vs. grade 3 p = 0.017; grade 2 vs. grade 3 p = 0.0437; grade 1 vs. grade 2&3 p = 0.021) and Nottingham Prognostic Index (NPI) (NPI-1 vs. NPI-3 p = 0.0614; NPI-2 vs. NPI-3 p = 0.0495). Transcript levels were associated with oestrogen receptor (ER) positivity (ER(-) vs. ER(+) p = 0.046). SABT1 expression was also significantly correlated with downstream regulated genes IL-4 and MAF-1 (Pearson's correlation coefficient r = 0.21 and r = 0.162) and SATB2 (r = 0.506). After a median follow up of 10 years, there was a trend for higher SATB1 expression to be associated with shorter overall survival (OS). Higher levels of SATB2 were also found in malignant compared to background tissue (p = 0.049). SATB2 expression increased with increasing tumour grade (grade 1 vs. grade 3 p = 0.035). SATB2 was associated with ER positivity (ER(-) vs. ER(+) p = 0.0283) within ductal carcinomas. Higher transcript levels showed a significant association with poorer OS (p = 0.0433).
Conclusion: SATB1 mRNA expression is significantly associated with poor prognostic parameters in breast cancer, including increasing tumour grade, TNM stage and NPI. SATB2 mRNA expression is significantly associated with increasing tumour grade and poorer OS. These results are consistent with the notion that SATB1 acts as a 'master genome organizer' in human breast carcinogenesis.
Figures
References
-
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. - PubMed
-
- Freiman RN, Tjian R. Regulating the regulators: Lysine modifications make their mark. Cell. 2003;112:11–17. - PubMed
-
- Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

