Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa
- PMID: 19645300
- PMCID: PMC2741316
- DOI: 10.1603/033.046.0428
Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa
Abstract
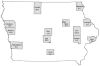
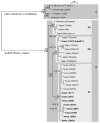
Adult mosquitoes (Diptera: Culicidae) were collected in 2007 and tested for specific viruses, including West Nile virus, as part of the ongoing arbovirus surveillance efforts in the state of Iowa. A subset of these mosquitoes (6,061 individuals in 340 pools) was further tested by reverse transcription-polymerase chain reaction (RT-PCR) using flavivirus universal primers. Of the 211 pools of Culex pipiens (L.) tested, 50 were positive. One of 51 pools of Culex tarsalis Coquillet was also positive. The flavivirus minimum infection rates (expressed as the number of positive mosquito pools per 1,000 mosquitoes tested) for Cx. pipiens and Cx. tarsalis were 10.3 and 1.2, respectively. Flavivirus RNA was not detected in Aedes triseriatus (Say) (52 pools), Culex erraticus (Dyar & Knab) (25 pools), or Culex territans Walker (one pool). Sequence analysis of all RT-PCR products revealed that the mosquitoes had been infected with Culex flavivirus (CxFV), an insect-specific virus previously isolated in Japan, Indonesia, Texas, Mexico, Guatemala and Trinidad. The complete genome of one isolate was sequenced, as were the envelope protein genes of eight other isolates. Phylogenetic analysis revealed that CxFV isolates from the United States (Iowa and Texas) are more closely related to CxFV isolates from Asia than those from Mexico, Guatemala, and Trinidad.
Figures

References
-
- Biggerstaff B. Software for mosquito surveillance. Centers for Disease Control and Prevention; Fort Collins, Colorado: 2006. cited 2009 April 3. http://www.cdc.gov/ncidod/dvbid/westnile/software.htm.
-
- Cammisa-Parks H, Cisar LA, Kane A, Stollar V. The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology. 1992;189:511–524. - PubMed
-
- Cook S, Holmes EC. A multigene analysis of the phylogenetic relationships among the flaviviruses (Family: Flaviviridae) and the evolution of vector transmission. Arch Virol. 2006;151:309–325. - PubMed
-
- Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol. 2006;87:735–748. - PubMed
-
- Crabtree MB, Sang RC, Stollar V, Dunster LM, Miller BR. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch Virol. 2003;148:1095–1118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

