Feedback regulation of NEUROG2 activity by MTGR1 is required for progression of neurogenesis
- PMID: 19646530
- PMCID: PMC2783839
- DOI: 10.1016/j.mcn.2009.07.011
Feedback regulation of NEUROG2 activity by MTGR1 is required for progression of neurogenesis
Abstract
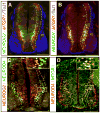
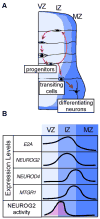
The sequential steps of neurogenesis are characterized by highly choreographed changes in transcription factor activity. In contrast to the well-studied mechanisms of transcription factor activation during neurogenesis, much less is understood regarding how such activity is terminated. We previously showed that MTGR1, a member of the MTG family of transcriptional repressors, is strongly induced by a proneural basic helix-loop-helix transcription factor, NEUROG2 in developing nervous system. In this study, we describe a novel feedback regulation of NEUROG2 activity by MTGR1. We show that MTGR1 physically interacts with NEUROG2 and represses transcriptional activity of NEUROG2. MTGR1 also prevents DNA binding of the NEUROG2/E47 complex. In addition, we provide evidence that proper termination of NEUROG2 activity by MTGR1 is necessary for normal progression of neurogenesis in the developing spinal cord. These results highlight the importance of feedback regulation of proneural gene activity in neurodevelopment.
Figures





References
-
- Amann JM, Chyla BJI, Ellis TC, Martinez A, Moore AC, Franklin JL, McGhee L, Meyers S, Ohm JE, Luce KS, Ouelette AJ, Washington MK, Thompson MA, King D, Gautam S, Coffey RJ, Whitehead RH, Hiebert SW. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Molecular and Cellular Biology. 2005;25:9576–9585. - PMC - PubMed
-
- Ausubel FM, editor. Current protocols in molecular biology. John Wiley & Sons, Inc; 1994.
-
- Cai L, Morrow EM, Cepko CL. Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development. 2000;127:3021–3030. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

