Protein occupancy landscape of a bacterial genome
- PMID: 19647521
- PMCID: PMC2763621
- DOI: 10.1016/j.molcel.2009.06.035
Protein occupancy landscape of a bacterial genome
Abstract
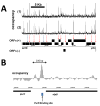
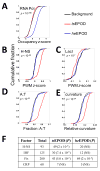
Protein-DNA interactions are fundamental to core biological processes, including transcription, DNA replication, and chromosomal organization. We have developed in vivo protein occupancy display (IPOD), a technology that reveals protein occupancy across an entire bacterial chromosome at the resolution of individual binding sites. Application to Escherichia coli reveals thousands of protein occupancy peaks, highly enriched within and in close proximity to noncoding regulatory regions. In addition, we discovered extensive (>1 kilobase) protein occupancy domains (EPODs), some of which are localized to highly expressed genes, enriched in RNA-polymerase occupancy. However, the majority are localized to transcriptionally silent loci dominated by conserved hypothetical ORFs. These regions are highly enriched in both predicted and experimentally determined binding sites of nucleoid proteins and exhibit extreme biophysical characteristics such as high intrinsic curvature. Our observations implicate these transcriptionally silent EPODs as the elusive organizing centers, long proposed to topologically isolate chromosomal domains.
Figures


Comment in
-
Can the protein occupancy landscape show the topologically isolated chromosomal domains in the E. coli genome?: An exciting prospect.Mol Cell. 2009 Aug 14;35(3):255-6. doi: 10.1016/j.molcel.2009.07.019. Mol Cell. 2009. PMID: 19683488
References
-
- Aiyar SE, McLeod SM, Ross W, Hirvonen CA, Thomas MS, Johnson RC, Gourse RL. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J Mol Biol. 2002;316:501–516. - PubMed
-
- Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. - PubMed
-
- Bendich AJ. The form of chromosomal DNA molecules in bacterial cells. Biochimie. 2001;83:177–186. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

