TCR repertoire and Foxp3 expression define functionally distinct subsets of CD4+ regulatory T cells
- PMID: 19648277
- PMCID: PMC2804979
- DOI: 10.4049/jimmunol.0900514
TCR repertoire and Foxp3 expression define functionally distinct subsets of CD4+ regulatory T cells
Abstract
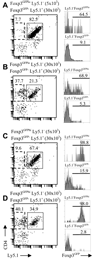
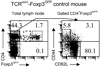
Despite extensive research efforts to characterize peripheral regulatory T (T(reg)) cells expressing transcription factor Foxp3, their subset complexity, phenotypic characteristics, TCR repertoire and Ag specificities remain ambiguous. In this study, we identify and define two subsets of peripheral T(reg) cells differing in Foxp3 expression level and TCR repertoires. T(reg) cells expressing a high level of Foxp3 and TCRs not used by naive CD4(+) T cells present a stable suppressor phenotype and dominate the peripheral T(reg) population in unmanipulated mice. The second T(reg) subset, expressing a lower level of Foxp3 and using TCRs shared with naive CD4(+) T cells constitutes a small fraction of all T(reg) cells in unmanipulated mice and enriches T(reg) population with the same Ag specificities as expressed by activated/effector T cells. This T(reg) subset undergoes extensive expansion during response to Ag when it becomes a major population of Ag-specific T(reg) cells. Thus, T(reg) cells expressing TCRs shared with naive CD4(+) T cells have a flexible phenotype and may down-regulate Foxp3 expression which may restore immune balance at the conclusion of immune response or convert these cells to effector T cells producing inflammatory cytokines.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures





References
-
- Sakaguchi S, Sakaguchi N, Shimizu J, Yamazaki S, Sakihama T, Itoh M, Kuniyasu Y, Nomura T, Toda M, Takahashi T. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol. Rev. 2001;182:18–32. - PubMed
-
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. - PubMed
-
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. - PubMed
-
- Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, von BH. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 2005;6:1219–1227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

