The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid
- PMID: 19648296
- PMCID: PMC2729616
- DOI: 10.1105/tpc.109.065821
The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid
Abstract
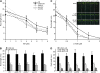
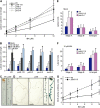
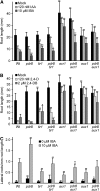
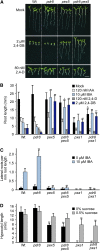
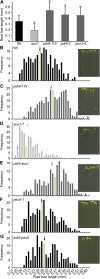
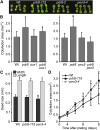
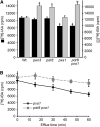
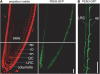
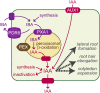
Plants have developed numerous mechanisms to store hormones in inactive but readily available states, enabling rapid responses to environmental changes. The phytohormone auxin has a number of storage precursors, including indole-3-butyric acid (IBA), which is apparently shortened to active indole-3-acetic acid (IAA) in peroxisomes by a process similar to fatty acid beta-oxidation. Whereas metabolism of auxin precursors is beginning to be understood, the biological significance of the various precursors is virtually unknown. We identified an Arabidopsis thaliana mutant that specifically restores IBA, but not IAA, responsiveness to auxin signaling mutants. This mutant is defective in PLEIOTROPIC DRUG RESISTANCE8 (PDR8)/PENETRATION3/ABCG36, a plasma membrane-localized ATP binding cassette transporter that has established roles in pathogen responses and cadmium transport. We found that pdr8 mutants display defects in efflux of the auxin precursor IBA and developmental defects in root hair and cotyledon expansion that reveal previously unknown roles for IBA-derived IAA in plant growth and development. Our results are consistent with the possibility that limiting accumulation of the IAA precursor IBA via PDR8-promoted efflux contributes to auxin homeostasis.
Figures

References
-
- Bailly, A., Sovero, V., Vincenzetti, V., Santelia, D., Bartnik, D., Koenig, B.W., Mancuso, S., Martinoia, E., and Geisler, M. (2008). Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 283 21817–21826. - PubMed
-
- Bandyopadhyay, A., et al. (2007). Interactions of PIN and PGP auxin transport mechanisms. Biochem. Soc. Trans. 35 137–141. - PubMed
-
- Bednarek, P., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. - PubMed
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

