Roles of trans and cis variation in yeast intraspecies evolution of gene expression
- PMID: 19648464
- PMCID: PMC2767097
- DOI: 10.1093/molbev/msp171
Roles of trans and cis variation in yeast intraspecies evolution of gene expression
Abstract
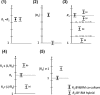
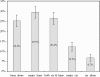
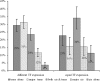
Both cis and trans mutations contribute to gene expression divergence within and between species. We used Saccharomyces cerevisiae as a model organism to estimate the relative contributions of cis and trans variations to the expression divergence between a laboratory (BY) and a wild (RM) strain of yeast. We examined whether genes regulated by a single transcription factor (TF; single input module, SIM genes) or genes regulated by multiple TFs (multiple input module, MIM genes) are more susceptible to trans variation. Because a SIM gene is regulated by a single immediate upstream TF, the chance for a change to occur in its trans-acting factors would, on average, be smaller than that for a MIM gene. We chose 232 genes that exhibited expression divergence between BY and RM to test this hypothesis. We examined the expression patterns of these genes in a BY-RM coculture system and in a BY-RM diploid hybrid. We found that trans variation is far more important than cis variation for expression divergence between the two strains. However, because in 75% of the genes studied, cis variation has significantly contributed to expression divergence, cis change also plays a significant role in intraspecific expression evolution. Interestingly, we found that the proportion of genes with diverged expression between BY and RM is larger for MIM genes than for SIM genes; in fact, the proportion tends to increase with the number of transcription factors that regulate the gene. Moreover, MIM genes are, on average, subject to stronger trans effects than SIM genes, though the difference between the two types of genes is not conspicuous.
Figures


References
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
-
- Mortimer RK, Romano P, Suzzi G, Polsinelli M. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast. 1994;10:1543–1552. - PubMed
-
- Osada N, Kohn MH, Wu CI. Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Mol Biol Evol. 2006;23:1585–1591. - PubMed
-
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

