Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models
- PMID: 19648916
- PMCID: PMC2762791
- DOI: 10.1038/nmeth.1358
Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models
Abstract
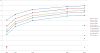
Metagenomics projects collect DNA from uncharacterized environments that may contain thousands of species per sample. One main challenge facing metagenomic analysis is phylogenetic classification of raw sequence reads into groups representing the same or similar taxa, a prerequisite for genome assembly and for analyzing the biological diversity of a sample. New sequencing technologies have made metagenomics easier, by making sequencing faster, and more difficult, by producing shorter reads than previous technologies. Classifying sequences from reads as short as 100 base pairs has until now been relatively inaccurate, requiring researchers to use older, long-read technologies. We present Phymm, a classifier for metagenomic data, that has been trained on 539 complete, curated genomes and can accurately classify reads as short as 100 base pairs, a substantial improvement over previous composition-based classification methods. We also describe how combining Phymm with sequence alignment algorithms improves accuracy.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

