Looking deeper: whole-mount confocal imaging of plant tissue for the accurate study of inner tissue layers
- PMID: 19649197
- PMCID: PMC2637507
- DOI: 10.4161/psb.4.2.7683
Looking deeper: whole-mount confocal imaging of plant tissue for the accurate study of inner tissue layers
Abstract
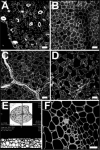
Building on previous work, we further developed a staining procedure that fluorescently labels plant cell walls and, when combined with confocal microscopy, allows visualization of plant cellular organisation in whole mounts to depths exceeding 200 microm. This technique can be combined with beta-glucuronidase histochemical activity assays, allowing the simultaneous study of gene expression. Images taken from stained samples can be used for three-dimensional tissue reconstruction. We used the technique to study Arabidopsis protophloem development. The phloem is one of the innermost tissues of a plant and therefore especially difficult to visualise. Furthermore, in general the technique will improve significantly the speed and accuracy with which any kind of plant tissue organisation can be studied, and it is not limited to the study of Arabidopsis tissues.
Keywords: confocal microscopy; gene expression; plant development; three-dimensional imaging; vasculature; whole-mount.
Figures
Comment on
-
High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis.Plant Cell. 2008 Jun;20(6):1494-503. doi: 10.1105/tpc.107.056069. Epub 2008 Jun 3. Plant Cell. 2008. PMID: 18523061 Free PMC article.
References
-
- Truernit E, Siemering KR, Hodge S, Grbic V, Haseloff J. A map of KNAT gene expression in the Arabidopsis root. Plant Mol Biol. 2006;60:1–20. - PubMed
-
- Noriega A, Cervantes E, Tocino A. Ethylene responses in Arabidopsis seedlings include the reduction of curvature values in the root cap. J Plant Physiol. 2008;165:960–966. - PubMed
-
- Moreno N, Bougourd S, Haseloff J, Feijo J. Imaging Plant Cells. In: Pawley J, editor. Handbook of Biological Confocal Microscopy. New York: Springer Science and Business Media; 2006. pp. 769–787.
Publication types
LinkOut - more resources
Full Text Sources
