Retroelements and their impact on genome evolution and functioning
- PMID: 19649766
- PMCID: PMC11115525
- DOI: 10.1007/s00018-009-0107-2
Retroelements and their impact on genome evolution and functioning
Abstract
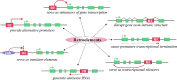
Retroelements comprise a considerable fraction of eukaryotic genomes. Since their initial discovery by Barbara McClintock in maize DNA, retroelements have been found in genomes of almost all organisms. First considered as a "junk DNA" or genomic parasites, they were shown to influence genome functioning and to promote genetic innovations. For this reason, they were suggested as an important creative force in the genome evolution and adaptation of an organism to altered environmental conditions. In this review, we summarize the up-to-date knowledge of different ways of retroelement involvement in structural and functional evolution of genes and genomes, as well as the mechanisms generated by cells to control their retrotransposition.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

