DNA-mediated redox signaling for transcriptional activation of SoxR
- PMID: 19651620
- PMCID: PMC2726364
- DOI: 10.1073/pnas.0906429106
DNA-mediated redox signaling for transcriptional activation of SoxR
Abstract
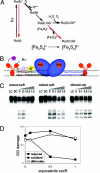
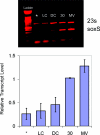
In enteric bacteria, the cellular response to oxidative stress is activated by oxidation of the iron-sulfur clusters in SoxR, which then induces transcription of soxS, turning on a battery of defense genes. Here we demonstrate both in vitro and in cells that activation of SoxR can occur in a DNA-mediated reaction with guanine radicals, an early genomic signal of oxidative stress, serving as the oxidant. SoxR in its reduced form is found to inhibit guanine damage by repairing guanine radicals. Moreover, cells treated with a DNA-binding photooxidant, which generates guanine radicals, promotes the expression of soxS. In vitro, this photooxidant, tethered to DNA 80 bp from the soxS promoter, induces transcription by activating SoxR upon irradiation. Thus, transcription can be activated from a distance through DNA-mediated charge transport. This chemistry offers a general strategy for DNA-mediated signaling of oxidative stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J Biol Chem. 1997;272:5082–5086. - PubMed
-
- Wu J, Dunham WR, Weiss B. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative stress response regulon in Escherichia coli. J Biol Chem. 1995;270:10323–10327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

