The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae
- PMID: 19652178
- PMCID: PMC2766306
- DOI: 10.1534/genetics.109.106229
The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae
Abstract
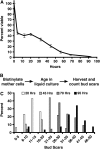
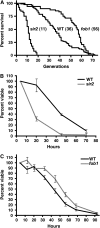
The replicative life span (RLS) of Saccharomyces cerevisiae has been established as a model for the genetic regulation of longevity despite the inherent difficulty of the RLS assay, which requires separation of mother and daughter cells by micromanipulation after every division. Here we present the mother enrichment program (MEP), an inducible genetic system in which mother cells maintain a normal RLS--a median of 36 generations in the diploid MEP strain--while the proliferative potential of daughter cells is eliminated. Thus, the viability of a population over time becomes a function of RLS, and it displays features of a survival curve such as changes in hazard rate with age. We show that viability of mother cells in liquid culture is regulated by SIR2 and FOB1, two opposing regulators of RLS in yeast. We demonstrate that viability curves of these short- and long-lived strains can be easily distinguished from wild type, using a colony formation assay. This provides a simplified screening method for identifying genetic or environmental factors that regulate RLS. Additionally, the MEP can provide a cohort of cells at any stage of their life span for the analysis of age-associated phenotypes. These capabilities effectively remove the hurdles presented by RLS analysis that have hindered S. cerevisiae aging studies since their inception 50 years ago.
Figures



References
-
- Ballou, C. E., 1982. Yeast cell wall and cell surface, pp. 335–360 in The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Chen, C., and R. Contreras, 2007. Identifying genes that extend life span using a high-throughput screening system. Methods Mol. Biol. 371: 237–248. - PubMed
-
- Colman-Lerner, A., T. E. Chin and R. Brent, 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107: 739–750. - PubMed
-
- Cost, G. J., and J. D. Boeke, 1996. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12: 939–941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

