Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana
- PMID: 19652183
- PMCID: PMC2766330
- DOI: 10.1534/genetics.109.104984
Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana
Abstract
Flowering time, a critical adaptive trait, is modulated by several environmental cues. These external signals converge on a small set of genes that in turn mediate the flowering response. Mutant analysis and subsequent molecular studies have revealed that one of these integrator genes, FLOWERING LOCUS T (FT), responds to photoperiod and temperature cues, two environmental parameters that greatly influence flowering time. As the central player in the transition to flowering, the protein coding sequence of FT and its function are highly conserved across species. Using QTL mapping with a new advanced intercross-recombinant inbred line (AI-RIL) population, we show that a QTL tightly linked to FT contributes to natural variation in the flowering response to the combined effects of photoperiod and ambient temperature. Using heterogeneous inbred families (HIF) and introgression lines, we fine map the QTL to a 6.7 kb fragment in the FT promoter. We confirm by quantitative complementation that FT has differential activity in the two parental strains. Further support for FT underlying the QTL comes from a new approach, quantitative knockdown with artificial microRNAs (amiRNAs). Consistent with the causal sequence polymorphism being in the promoter, we find that the QTL affects FT expression. Taken together, these results indicate that allelic variation at pathway integrator genes such as FT can underlie phenotypic variability and that this may be achieved through cis-regulatory changes.
Figures

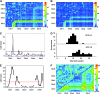


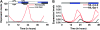
References
-
- Abe, M., Y. Kobayashi, S. Yamamoto, Y. Daimon, A. Yamaguchi et al., 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

