Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes
- PMID: 19653904
- PMCID: PMC2907695
- DOI: 10.1186/1471-2164-10-354
Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes
Abstract
Background: The recently described Type VI Secretion System (T6SS) represents a new paradigm of protein secretion in bacteria. A number of bioinformatic studies have been conducted to identify T6SS gene clusters in the available bacterial genome sequences. According to these studies, Salmonella harbors a unique T6SS encoded in the Salmonella Pathogenicity Island 6 (SPI-6). Since these studies only considered few Salmonella genomes, the present work aimed to identify novel T6SS loci by in silico analysis of every genome sequence of Salmonella available.
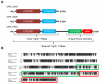
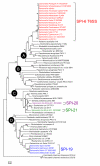
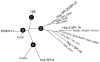
Results: The analysis of sequencing data from 44 completed or in progress Salmonella genome projects allowed the identification of 3 novel T6SS loci. These clusters are located in differentially-distributed genomic islands we designated SPI-19, SPI-20 and SPI-21, respectively. SPI-19 was identified in a subset of S. enterica serotypes including Dublin, Weltevreden, Agona, Gallinarum and Enteritidis. In the later, an internal deletion eliminated most of the island. On the other hand, SPI-20 and SPI-21 were restricted to S. enterica subspecies arizonae (IIIa) serotype 62:z4,z23:-. Remarkably, SPI-21 encodes a VgrG protein containing a C-terminal extension similar to S-type pyocins of Pseudomonas aeruginosa. This is not only the first evolved VgrG described in Salmonella, but also the first evolved VgrG including a pyocin domain described so far in the literature. In addition, the data indicate that SPI-6 T6SS is widely distributed in S. enterica and absent in serotypes Enteritidis, Gallinarum, Agona, Javiana, Paratyphi B, Virchow, IIIa 62:z4,z23:- and IIIb 61:1,v:1,5,(7). Interestingly, while some serotypes harbor multiple T6SS (Dublin, Weltvreden and IIIa 62:z4,z23:-) others do not encode for any (Enteritidis, Paratyphi B, Javiana, Virchow and IIIb 61:1,v:1,5,(7)). Comparative and phylogenetic analyses indicate that the 4 T6SS loci in Salmonella have a distinct evolutionary history. Finally, we identified an orphan Hcp-like protein containing the Hcp/COG3157 domain linked to a C-terminal extension. We propose to designate this and related proteins as "evolved Hcps".
Conclusion: Altogether, our data suggest that (i) the Salmonella T6SS loci were acquired by independent lateral transfer events and (ii) evolved to contribute in the adaptation of the serotypes to different lifestyles and environments, including animal hosts. Notably, the presence of an evolved VgrG protein related to pyocins suggests a novel role for T6SS in bacterial killing. Future studies on the roles of the identified T6SS loci will expand our knowledge on Salmonella pathogenesis and host specificity.
Figures





References
-
- Das S, Chaudhuri K. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 2003;3(3):287–300. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

