Differences in conformational dynamics of [Pt3(HPTAB)]6+-DNA adducts with various cross-linking modes
- PMID: 19654239
- PMCID: PMC2761282
- DOI: 10.1093/nar/gkp618
Differences in conformational dynamics of [Pt3(HPTAB)]6+-DNA adducts with various cross-linking modes
Abstract
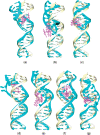
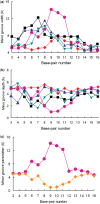
We present here molecular dynamics simulations and DNA conformational dynamics for a series of trinuclear platinum [Pt(3)(HPTAB)](6+)-DNA adducts [HPTAB = N,N,N',N',N'',N''-hexakis (2-pyridyl-methyl)-1,3,5-tris(aminomethyl) benzene], including three types of bifunctional crosslinks and four types of trifunctional crosslinks. Our simulation results reveal that binding of the trinuclear platinum compound to a DNA duplex induces the duplex unwinding in the vicinity of the platination sites, and causes the DNA to bend toward the major groove. As a consequence, this produces a DNA molecule whose minor groove is more widened and shallow compared to that of an undamaged bare-DNA molecule. Notably, for trifunctional crosslinks, we have observed extensive DNA conformational distortions, which is rarely seen for normal platinum-DNA adducts. Our findings, in this study, thus provide further support for the idea that platinum compounds with trifunctional intra-strand or long-range-inter-strand cross-linking modes can generate larger DNA conformational distortions than other types of cross-linking modes.
Figures




Similar articles
-
DNA cross-linking patterns induced by an antitumor-active trinuclear platinum complex and comparison with its dinuclear analogue.Chemistry. 2009;15(21):5245-53. doi: 10.1002/chem.200900217. Chemistry. 2009. PMID: 19350599
-
Unique DNA binding mode of antitumor trinuclear tridentate platinum(II) compound.Mol Pharm. 2011 Dec 5;8(6):2368-78. doi: 10.1021/mp200298g. Epub 2011 Nov 16. Mol Pharm. 2011. PMID: 22050363
-
Long range 1,4 and 1,6-interstrand cross-links formed by a trinuclear platinum complex. Minor groove preassociation affects kinetics and mechanism of cross-link formation as well as adduct structure.J Am Chem Soc. 2004 Feb 25;126(7):2166-80. doi: 10.1021/ja036105u. J Am Chem Soc. 2004. PMID: 14971952
-
NMR spectroscopy of anticancer platinum drugs.Anticancer Agents Med Chem. 2007 Jan;7(1):35-54. doi: 10.2174/187152007779313982. Anticancer Agents Med Chem. 2007. PMID: 17266504 Review.
-
Interstrand cross-links of cisplatin induce striking distortions in DNA.J Inorg Biochem. 1999 Oct;77(1-2):23-9. doi: 10.1016/s0162-0134(99)00148-8. J Inorg Biochem. 1999. PMID: 10626349 Review.
Cited by
-
Investigations of the binding of [Pt2(DTBPA)Cl2](II) and [Pt2(TPXA)Cl2](II) to DNA via various cross-linking modes.Int J Mol Sci. 2013 Sep 26;14(10):19556-86. doi: 10.3390/ijms141019556. Int J Mol Sci. 2013. PMID: 24077126 Free PMC article.
-
Disturbance of DNA conformation by the binding of testosterone-based platinum drugs via groove-face and intercalative interactions: a molecular dynamics simulation study.BMC Struct Biol. 2013 Mar 22;13:4. doi: 10.1186/1472-6807-13-4. BMC Struct Biol. 2013. PMID: 23517640 Free PMC article.
References
-
- Hegmans A, Berners-Price SJ, Davies MS, Thomas DS, Humphreys AS, Farrell N. Long range 1,4 and 1,6-interstrand cross-links formed by a trinuclear platinum complex. Minor groove preassociation affects kinetics and mechanism of cross-link formation as well as adduct structure. J. Am. Chem. Soc. 2004;126:2166–2180. - PubMed
-
- Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007;107:1387–1407. - PubMed
-
- Berners-Price SJ, Davies MS, Cox JW, Thomas DS, Farrell N. Competitive reactions of interstrand and intrastrand DNA-Pt adducts: a dinuclear-platinum complex preferentially forms a 1,4-interstrand cross-link rather than a 1,2 intrastrand cross-link on binding to a GG 14-mer duplex. Chem. Eur. J. 2003;9:713–725. - PubMed
-
- Kasparkova J, Zehnulova J, Farrell N, Brabec V. DNA interstrand cross-links of the novel antitumor trinuclear platinum complex BBR3464. conformation, recognition by high mobility group domain proteins, and nucleotide excision repair. J. Biol. Chem. 2002;277:48076–48086. - PubMed
-
- Wong E, Giandomenico CM. Current status of platinum-based antitumor drugs. Chem. Rev. 1999;99:2451–2466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

