The derivation of diagnostic markers of chronic myeloid leukemia progression from microarray data
- PMID: 19654405
- PMCID: PMC2759651
- DOI: 10.1182/blood-2009-03-212969
The derivation of diagnostic markers of chronic myeloid leukemia progression from microarray data
Abstract
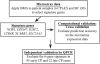
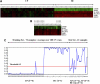
Currently, limited molecular markers exist that can determine where in the spectrum of chronic myeloid leukemia (CML) progression an individual patient falls at diagnosis. Gene expression profiles can predict disease and prognosis, but most widely used microarray analytical methods yield lengthy gene candidate lists that are difficult to apply clinically. Consequently, we applied a probabilistic method called Bayesian model averaging (BMA) to a large CML microarray dataset. BMA, a supervised method, considers multiple genes simultaneously and identifies small gene sets. BMA identified 6 genes (NOB1, DDX47, IGSF2, LTB4R, SCARB1, and SLC25A3) that discriminated chronic phase (CP) from blast crisis (BC) CML. In CML, phase labels divide disease progression into discrete states. BMA, however, produces posterior probabilities between 0 and 1 and predicts patients in "intermediate" stages. In validation studies of 88 patients, the 6-gene signature discriminated early CP from late CP, accelerated phase, and BC. This distinction between early and late CP is not possible with current classifications, which are based on known duration of disease. BMA is a powerful tool for developing diagnostic tests from microarray data. Because therapeutic outcomes are so closely tied to disease phase, these probabilities can be used to determine a risk-based treatment strategy at diagnosis.
Figures


References
-
- Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. - PubMed
-
- Faderl S, Talpaz M, Estrov Z, O'Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341(3):164–172. - PubMed
-
- Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–1037. - PubMed
-
- Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. - PubMed
-
- Sawyers CL, Hochhaus A, Feldman E, et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99(10):3530–3539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD054511/HD/NICHD NIH HHS/United States
- U54 AI057141/AI/NIAID NIH HHS/United States
- CA18029/CA/NCI NIH HHS/United States
- K25CA106988/CA/NCI NIH HHS/United States
- P01 CA018029/CA/NCI NIH HHS/United States
- R24RR021863-01A1/RR/NCRR NIH HHS/United States
- R01HDO54511-01A1/PHS HHS/United States
- R01 GM084163/GM/NIGMS NIH HHS/United States
- U54AI057141/AI/NIAID NIH HHS/United States
- UL1 RR025014/RR/NCRR NIH HHS/United States
- R01 DE012212/DE/NIDCR NIH HHS/United States
- UL1RR025014-01/RR/NCRR NIH HHS/United States
- R01GM084163-01A1/GM/NIGMS NIH HHS/United States
- R24 HD042828/HD/NICHD NIH HHS/United States
- K25 CA106988/CA/NCI NIH HHS/United States
- P50HL073996/HL/NHLBI NIH HHS/United States
- R01 CA140371/CA/NCI NIH HHS/United States
- R24 RR021863/RR/NCRR NIH HHS/United States
- R01DE012212-06/DE/NIDCR NIH HHS/United States
- P50 HL073996/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

