Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology
- PMID: 19654876
- PMCID: PMC2716536
- DOI: 10.1371/journal.pone.0006524
Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology
Abstract
Background: The dissection of complex traits of economic importance to the pig industry requires the availability of a significant number of genetic markers, such as single nucleotide polymorphisms (SNPs). This study was conducted to discover several hundreds of thousands of porcine SNPs using next generation sequencing technologies and use these SNPs, as well as others from different public sources, to design a high-density SNP genotyping assay.
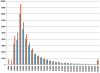
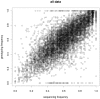
Methodology/principal findings: A total of 19 reduced representation libraries derived from four swine breeds (Duroc, Landrace, Large White, Pietrain) and a Wild Boar population and three restriction enzymes (AluI, HaeIII and MspI) were sequenced using Illumina's Genome Analyzer (GA). The SNP discovery effort resulted in the de novo identification of over 372K SNPs. More than 549K SNPs were used to design the Illumina Porcine 60K+SNP iSelect Beadchip, now commercially available as the PorcineSNP60. A total of 64,232 SNPs were included on the Beadchip. Results from genotyping the 158 individuals used for sequencing showed a high overall SNP call rate (97.5%). Of the 62,621 loci that could be reliably scored, 58,994 were polymorphic yielding a SNP conversion success rate of 94%. The average minor allele frequency (MAF) for all scorable SNPs was 0.274.
Conclusions/significance: Overall, the results of this study indicate the utility of using next generation sequencing technologies to identify large numbers of reliable SNPs. In addition, the validation of the PorcineSNP60 Beadchip demonstrated that the assay is an excellent tool that will likely be used in a variety of future studies in pigs.
Conflict of interest statement
Figures



References
-
- Hu Z-L, Reecy JM. Animal QTLdb: beyond a repository. A public platform for QTL comparisons and integration with diverse types of structural genomic information. Mamm Genom. 2007;18(1):1–4. - PubMed
-
- Jungerius BJ, Gu J, Crooijmans RPMA, Poel JJ, Groenen MAM, et al. Estimation of the extent of linkage disequilibrium in seven regions of the porcine genome. Anim Biotechnol. 2005;16:41–54. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/E010520/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05191130/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- EGA16307/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- EGA16307/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E010520/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/05478/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/R/00000685/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F021372/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/05478/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

