Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content
- PMID: 19656368
- PMCID: PMC2907686
- DOI: 10.1186/1471-2164-10-358
Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content
Abstract
Background: Streptococcus mutans is the major pathogen of dental caries, and it occasionally causes infective endocarditis. While the pathogenicity of this species is distinct from other human pathogenic streptococci, the species-specific evolution of the genus Streptococcus and its genomic diversity are poorly understood.
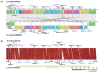
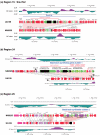
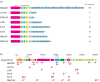
Results: We have sequenced the complete genome of S. mutans serotype c strain NN2025, and compared it with the genome of UA159. The NN2025 genome is composed of 2,013,587 bp, and the two strains show highly conserved core-genome. However, comparison of the two S. mutans strains showed a large genomic inversion across the replication axis producing an X-shaped symmetrical DNA dot plot. This phenomenon was also observed between other streptococcal species, indicating that streptococcal genetic rearrangements across the replication axis play an important role in Streptococcus genetic shuffling. We further confirmed the genomic diversity among 95 clinical isolates using long-PCR analysis. Genomic diversity in S. mutans appears to occur frequently between insertion sequence (IS) elements and transposons, and these diversity regions consist of restriction/modification systems, antimicrobial peptide synthesis systems, and transporters. S. mutans may preferentially reject the phage infection by clustered regularly interspaced short palindromic repeats (CRISPRs). In particular, the CRISPR-2 region, which is highly divergent between strains, in NN2025 has long repeated spacer sequences corresponding to the streptococcal phage genome.
Conclusion: These observations suggest that S. mutans strains evolve through chromosomal shuffling and that phage infection is not needed for gene acquisition. In contrast, S. pyogenes tolerates phage infection for acquisition of virulence determinants for niche adaptation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

