Multi-level learning: improving the prediction of protein, domain and residue interactions by allowing information flow between levels
- PMID: 19656385
- PMCID: PMC2734556
- DOI: 10.1186/1471-2105-10-241
Multi-level learning: improving the prediction of protein, domain and residue interactions by allowing information flow between levels
Abstract
Background: Proteins interact through specific binding interfaces that contain many residues in domains. Protein interactions thus occur on three different levels of a concept hierarchy: whole-proteins, domains, and residues. Each level offers a distinct and complementary set of features for computationally predicting interactions, including functional genomic features of whole proteins, evolutionary features of domain families and physical-chemical features of individual residues. The predictions at each level could benefit from using the features at all three levels. However, it is not trivial as the features are provided at different granularity.
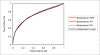
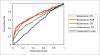
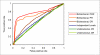
Results: To link up the predictions at the three levels, we propose a multi-level machine-learning framework that allows for explicit information flow between the levels. We demonstrate, using representative yeast interaction networks, that our algorithm is able to utilize complementary feature sets to make more accurate predictions at the three levels than when the three problems are approached independently. To facilitate application of our multi-level learning framework, we discuss three key aspects of multi-level learning and the corresponding design choices that we have made in the implementation of a concrete learning algorithm. 1) Architecture of information flow: we show the greater flexibility of bidirectional flow over independent levels and unidirectional flow; 2) Coupling mechanism of the different levels: We show how this can be accomplished via augmenting the training sets at each level, and discuss the prevention of error propagation between different levels by means of soft coupling; 3) Sparseness of data: We show that the multi-level framework compounds data sparsity issues, and discuss how this can be dealt with by building local models in information-rich parts of the data. Our proof-of-concept learning algorithm demonstrates the advantage of combining levels, and opens up opportunities for further research.
Availability: The software and a readme file can be downloaded at http://networks.gersteinlab.org/mll. The programs are written in Java, and can be run on any platform with Java 1.4 or higher and Apache Ant 1.7.0 or higher installed. The software can be used without a license.
Figures

References
-
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a Protein-Protein Interaction Map of the Budding Yeast: A Comprehensive System to Examine Two-Hybrid Interactions in All Possible Combinations between the Yeast Proteins. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. - DOI - PMC - PubMed
-
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a Proteome-scale Map of the Human Protein-Protein Interaction Network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. - DOI - PubMed
-
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell. 2005;122:967–968. doi: 10.1016/j.cell.2005.08.029. - DOI - PubMed
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A Comprehensive Analysis of Protein-Protein Interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. - DOI - PubMed
-
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome Survey Reveals Modularity of the Yeast Cell Machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

