Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis
- PMID: 19656490
- PMCID: PMC2745341
- DOI: 10.1016/j.cmet.2009.06.012
Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis
Abstract
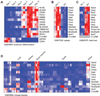
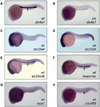
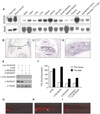
Heme biosynthesis consists of a series of eight enzymatic reactions that originate in mitochondria and continue in the cytosol before returning to mitochondria. Although these core enzymes are well studied, additional mitochondrial transporters and regulatory factors are predicted to be required. To discover such unknown components, we utilized a large-scale computational screen to identify mitochondrial proteins whose transcripts consistently coexpress with the core machinery of heme biosynthesis. We identified SLC25A39, SLC22A4, and TMEM14C, which are putative mitochondrial transporters, as well as C1orf69 and ISCA1, which are iron-sulfur cluster proteins. Targeted knockdowns of all five genes in zebrafish resulted in profound anemia without impacting erythroid lineage specification. Moreover, silencing of Slc25a39 in murine erythroleukemia cells impaired iron incorporation into protoporphyrin IX, and vertebrate Slc25a39 complemented an iron homeostasis defect in the orthologous yeast mtm1Delta deletion mutant. Our results advance the molecular understanding of heme biosynthesis and offer promising candidate genes for inherited anemias.
Figures



Comment in
-
Tangled up in red: intertwining of the heme and iron-sulfur cluster biogenesis pathways.Cell Metab. 2009 Aug;10(2):80-1. doi: 10.1016/j.cmet.2009.07.007. Cell Metab. 2009. PMID: 19656484
References
-
- Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763:723–736. - PubMed
-
- Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum Mol Genet. 1999;8:743–749. - PubMed
-
- Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. - PubMed
-
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01DK052380/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01GM077465/GM/NIGMS NIH HHS/United States
- P01HL032262/HL/NHLBI NIH HHS/United States
- R01DK070838/DK/NIDDK NIH HHS/United States
- R01 ES008996/ES/NIEHS NIH HHS/United States
- P30 ES003819/ES/NIEHS NIH HHS/United States
- P01 HL032262/HL/NHLBI NIH HHS/United States
- R01 DK070838/DK/NIDDK NIH HHS/United States
- R01ES08996/ES/NIEHS NIH HHS/United States
- R01 GM077465/GM/NIGMS NIH HHS/United States
- R01 DK052380/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

