Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain
- PMID: 19656744
- PMCID: PMC2748156
- DOI: 10.1016/j.dnarep.2009.07.003
Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain
Abstract
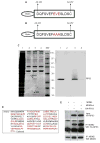
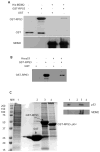
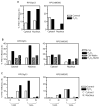
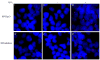
The p53 protein responds to cellular stress and regulates genes involved in cell cycle, apoptosis, and DNA repair. Under normal conditions, p53 levels are kept low through MDM2-mediated ubiquitination and proteosomal degradation. In search for novel proteins that participate in this regulatory loop, we performed an MDM2 peptide pull-down assay and mass spectrometry to screen for potential interacting partners of MDM2. We identified ribosomal protein S3 (RPS3), whose interaction with MDM2, and notably p53, was further established by His and GST pull-down assays, fluorescence resonance energy transfer and an in situ proximity ligation assay. Additionally, in cells exposed to oxidative stress, p53 levels increased slightly over 24h, whereas MDM2 levels declined after 6h exposure, but rose over the next 18h of exposure. Conversely, in cells exposed to oxidative stress and harboring siRNA to knockdown RPS3 expression, decreased p53 levels and loss of the E3 ubiquitin ligase domain possessed by MDM2 were observed. DNA pull-down assays using a 7,8-dihydro-8-oxoguanine duplex oligonucleotide as a substrate found that RPS3 acted as a scaffold for the additional binding of MDM2 and p53, suggesting that RPS3 interacts with important proteins involved in maintaining genomic integrity.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Coutts AS, Adams CJ, La NB. Thangue p53 ubiquitination by Mdm2: A never ending tail? DNA Repair (Amst) 2009;8:483–490. - PubMed
-
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. - PubMed
-
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. - PubMed
-
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. - PubMed
-
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

