Activation of MDA5 requires higher-order RNA structures generated during virus infection
- PMID: 19656871
- PMCID: PMC2753146
- DOI: 10.1128/JVI.00770-09
Activation of MDA5 requires higher-order RNA structures generated during virus infection
Abstract
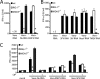
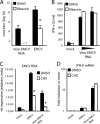
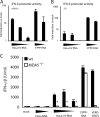
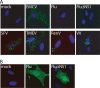
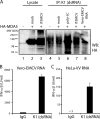
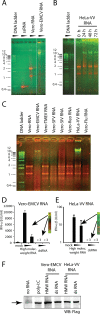
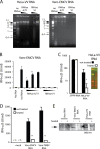
Recognition of virus presence via RIG-I (retinoic acid inducible gene I) and/or MDA5 (melanoma differentiation-associated protein 5) initiates a signaling cascade that culminates in transcription of innate response genes such as those encoding the alpha/beta interferon (IFN-alpha/beta) cytokines. It is generally assumed that MDA5 is activated by long molecules of double-stranded RNA (dsRNA) produced by annealing of complementary RNAs generated during viral infection. Here, we used an antibody to dsRNA to show that the presence of immunoreactivity in virus-infected cells does indeed correlate with the ability of RNA extracted from these cells to activate MDA5. Furthermore, RNA from cells infected with encephalomyocarditis virus or with vaccinia virus and precipitated with the anti-dsRNA antibody can bind to MDA5 and induce MDA5-dependent IFN-alpha/beta production upon transfection into indicator cells. However, a prominent band of dsRNA apparent in cells infected with either virus does not stimulate IFN-alpha/beta production. Instead, stimulatory activity resides in higher-order structured RNA that contains single-stranded RNA and dsRNA. These results suggest that MDA5 activation requires an RNA web rather than simply long molecules of dsRNA.
Figures

Similar articles
-
Inhibition of antiviral innate immunity by birnavirus VP3 protein via blockage of viral double-stranded RNA binding to the host cytoplasmic RNA detector MDA5.J Virol. 2014 Oct;88(19):11154-65. doi: 10.1128/JVI.01115-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031338 Free PMC article.
-
MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells.Cell Rep. 2012 Nov 29;2(5):1187-96. doi: 10.1016/j.celrep.2012.10.005. Epub 2012 Nov 8. Cell Rep. 2012. PMID: 23142662 Free PMC article.
-
Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses.Nature. 2006 May 4;441(7089):101-5. doi: 10.1038/nature04734. Epub 2006 Apr 9. Nature. 2006. PMID: 16625202
-
MDA5-filament, dynamics and disease.Curr Opin Virol. 2015 Jun;12:20-5. doi: 10.1016/j.coviro.2015.01.011. Epub 2015 Feb 9. Curr Opin Virol. 2015. PMID: 25676875 Free PMC article. Review.
-
LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review.
Cited by
-
A virological view of innate immune recognition.Annu Rev Microbiol. 2012;66:177-96. doi: 10.1146/annurev-micro-092611-150203. Annu Rev Microbiol. 2012. PMID: 22994491 Free PMC article. Review.
-
TLR3 signaling is either protective or pathogenic for the development of Theiler's virus-induced demyelinating disease depending on the time of viral infection.J Neuroinflammation. 2011 Dec 21;8:178. doi: 10.1186/1742-2094-8-178. J Neuroinflammation. 2011. PMID: 22189096 Free PMC article.
-
Defining the subcellular sites of innate immune signal transduction.Trends Immunol. 2012 Sep;33(9):442-8. doi: 10.1016/j.it.2012.06.005. Epub 2012 Jul 18. Trends Immunol. 2012. PMID: 22817912 Free PMC article. Review.
-
Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response.Adv Virus Res. 2016;96:219-243. doi: 10.1016/bs.aivir.2016.08.006. Epub 2016 Sep 9. Adv Virus Res. 2016. PMID: 27712625 Free PMC article. Review.
-
The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication.Nucleic Acids Res. 2014 Apr;42(7):4527-45. doi: 10.1093/nar/gku075. Epub 2014 Jan 26. Nucleic Acids Res. 2014. PMID: 24470146 Free PMC article.
References
-
- Colby, C., and P. H. Duesberg. 1969. Double-stranded RNA in vaccinia virus infected cells. Nature 222:940-944. - PubMed
-
- Declais, A. C., and D. M. Lilley. 2008. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opin. Struct. Biol. 18:86-95. - PubMed
-
- Delaloye, J., T. Roger, Q. G. Steiner-Tardivel, D. Le Roy, M. Knaup Reymond, S. Akira, V. Petrilli, C. E. Gomez, B. Perdiguero, J. Tschopp, G. Pantaleo, M. Esteban, and T. Calandra. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480. - PMC - PubMed
-
- Grunberg-Manago, M., P. J. Ortiz, and S. Ochoa. 1956. Enzymic synthesis of polynucleotides. I. Polynucleotide phosphorylase of azotobacter vinelandii. Biochim. Biophys. Acta 20:269-285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

