The molecular characterization and clinical management of multiple myeloma in the post-genome era
- PMID: 19657360
- PMCID: PMC3686133
- DOI: 10.1038/leu.2009.160
The molecular characterization and clinical management of multiple myeloma in the post-genome era
Abstract
Cancer-causing mutations disrupt coordinated, precise programs of gene expression that govern cell growth and differentiation. Microarray-based gene-expression profiling (GEP) is a powerful tool to globally analyze these changes to study cancer biology and clinical behavior. Despite overwhelming genomic chaos in multiple myeloma (MM), expression patterns within tumor samples are remarkably stable and reproducible. Unique expression patterns associated with recurrent chromosomal translocations and ploidy changes defined molecular classes with differing clinical features and outcomes. Combined molecular techniques also dissected two distinct, reproducible forms of hyperdiploid disease and have molecularly defined MM with high risk for poor clinical outcome. GEP is now used to risk-stratify patients with newly diagnosed MM. Groups with high-risk features are evident in all GEP-defined MM classes, and GEP studies of serial samples showed that risk increases over time, with relapsed disease showing dramatic GEP shifts toward a signature of poor outcomes. This suggests a common mechanism of disease evolution and potentially reflects preferential expansion of therapy-resistant cells. Correlating GEP-defined disease class and risk with outcomes of therapeutic regimens reveals class-specific benefits for individual agents, as well as mechanistic insights into drug sensitivity and resistance. Here, we review modern genomics contributions to understanding MM pathogenesis, prognosis, and therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
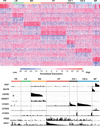
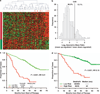
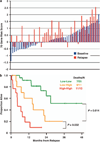
References
-
- Barlogie B, Shaughnessy J, Epstein J, Sanderson R, Anaissie E, Walker R, et al. Plasma cell myeloma. In: Lichtman MA, Beutler E, Kaushansky K, Kipps TJ, Seligsohn U, Prchal J, editors. Williams Hematology. 7 edn. New York: McGraw-Hill Professional; 2005. pp. 1501–1533.
-
- Ribatti D, Nico B, Vacca A. Importance of the bone marrow microenvironment in inducing the angiogenic response in multiple myeloma. Oncogene. 2006;25:4257–4266. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. - PubMed
-
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

