Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production
- PMID: 19659934
- PMCID: PMC4409829
- DOI: 10.1111/j.1365-2958.2009.06795.x
Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production
Abstract
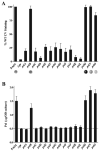
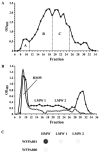
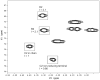
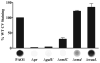
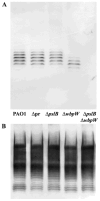
Exopolysaccharides contribute significantly to attachment and biofilm formation in the opportunisitc pathogen Pseudomonas aeruginosa. The Psl polysaccharide, which is synthesized by the polysaccharide synthesis locus (psl), is required for biofilm formation in non-mucoid strains that do not rely on alginate as the principal biofilm polysaccharide. In-frame deletion and complementation studies of individual psl genes revealed that 11 psl genes, pslACDEFGHIJKL, are required for Psl production and surface attachment. We also present the first structural analysis of the psl-dependent polysaccharide, which consists of a repeating pentasaccharide containing d-mannose, d-glucose and l-rhamnose: [See text]. In addition, we identified the sugar nucleotide precursors involved in Psl generation and demonstrated the requirement for GDP-d-mannose, UDP-d-glucose and dTDP-l-rhamnose in Psl production and surface attachment. Finally, genetic analyses revealed that wbpW restored Psl production in a pslB mutant and pslB promoted A-band LPS synthesis in a wbpW mutant, indicating functional redundancy and overlapping roles for these two enzymes. The structural and genetic data presented here provide a basis for further investigation of the Psl proteins and potential roles for Psl in the biology and pathogenesis of P. aeruginosa.
Figures


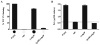

References
-
- Abeyrathne PD, Lam JS. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol Microbiol. 2007;65:1345–1359. - PubMed
-
- Bernatchez S, Szymanski CM, Ishiyama N, Li J, Jarrell HC, Lau PC, Berghuis AM, Young NM, Wakarchuk WW. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem. 2005;280:4792–4802. - PubMed
-
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. - PubMed
-
- Bystrova OV, Knirel YA, Lindner B, Kocharova NA, Kondakova AN, Zahringer U, Pier GB. Structures of the core oligosaccharide and O-units in the R- and SR-type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O-serogroups. FEMS Immunol Med Microbiol. 2006;46:85–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

