Phylogeny and mitochondrial gene order variation in Lophotrochozoa in the light of new mitogenomic data from Nemertea
- PMID: 19660126
- PMCID: PMC2728741
- DOI: 10.1186/1471-2164-10-364
Phylogeny and mitochondrial gene order variation in Lophotrochozoa in the light of new mitogenomic data from Nemertea
Abstract
Background: The new animal phylogeny established several taxa which were not identified by morphological analyses, most prominently the Ecdysozoa (arthropods, roundworms, priapulids and others) and Lophotrochozoa (molluscs, annelids, brachiopods and others). Lophotrochozoan interrelationships are under discussion, e.g. regarding the position of Nemertea (ribbon worms), which were discussed to be sister group to e.g. Mollusca, Brachiozoa or Platyhelminthes. Mitochondrial genomes contributed well with sequence data and gene order characters to the deep metazoan phylogeny debate.
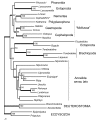
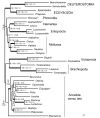
Results: In this study we present the first complete mitochondrial genome record for a member of the Nemertea, Lineus viridis. Except two trnP and trnT, all genes are located on the same strand. While gene order is most similar to that of the brachiopod Terebratulina retusa, sequence based analyses of mitochondrial genes place nemerteans close to molluscs, phoronids and entoprocts without clear preference for one of these taxa as sister group.
Conclusion: Almost all recent analyses with large datasets show good support for a taxon comprising Annelida, Mollusca, Brachiopoda, Phoronida and Nemertea. But the relationships among these taxa vary between different studies. The analysis of gene order differences gives evidence for a multiple independent occurrence of a large inversion in the mitochondrial genome of Lophotrochozoa and a re-inversion of the same part in gastropods. We hypothesize that some regions of the genome have a higher chance for intramolecular recombination than others and gene order data have to be analysed carefully to detect convergent rearrangement events.
Figures




References
-
- Halanych KM. The new view of animal phylogeny. Annu Rev Ecol Syst. 2004;35:229–256. doi: 10.1146/annurev.ecolsys.35.112202.130124. - DOI
-
- Mallatt J, Winchell CJ. Testing the new animal phylogeny: First use of combined large-subunit and small-subunit rRNA gene sequences to classify the protostomes. Mol Biol Evol. 2002;19:289–301. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

