Correlations between RNA and protein expression profiles in 23 human cell lines
- PMID: 19660143
- PMCID: PMC2728742
- DOI: 10.1186/1471-2164-10-365
Correlations between RNA and protein expression profiles in 23 human cell lines
Abstract
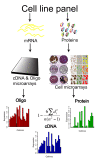
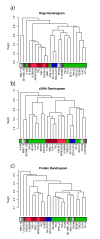
Background: The Central Dogma of biology holds, in famously simplified terms, that DNA makes RNA makes proteins, but there is considerable uncertainty regarding the general, genome-wide correlation between levels of RNA and corresponding proteins. Therefore, to assess degrees of this correlation we compared the RNA profiles (determined using both cDNA- and oligo-based microarrays) and protein profiles (determined immunohistochemically in tissue microarrays) of 1066 gene products in 23 human cell lines.
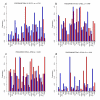
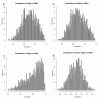
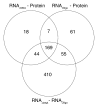
Results: A high mean correlation coefficient (0.52) was obtained from the pairwise comparison of RNA levels determined by the two platforms. Significant correlations, with correlation coefficients exceeding 0.445, between protein and RNA levels were also obtained for a third of the specific gene products. However, the correlation coefficients between levels of RNA and protein products of specific genes varied widely, and the mean correlations between the protein and corresponding RNA levels determined using the cDNA- and oligo-based microarrays were 0.25 and 0.20, respectively.
Conclusion: Significant correlations were found in one third of the examined RNA species and corresponding proteins. These results suggest that RNA profiling might provide indirect support to antibodies' specificity, since whenever a evident correlation between the RNA and protein profiles exists, this can sustain that the antibodies used in the immunoassay recognized their cognate antigens.
Figures
References
-
- Asplund A, Gry Bjorklund M, Sundquist C, Stromberg S, Edlund K, Ostman A, Nilsson P, Ponten F, Lundeberg J. Expression profiling of microdissected cell populations selected from basal cells in normal epidermis and basal cell carcinoma. The British journal of dermatology. 2008;158:527–538. doi: 10.1111/j.1365-2133.2007.08418.x. - DOI - PubMed
-
- Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, Cornelison R, Goldberger N, Elkahloun AG, Willi N, Koivisto P, et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer research. 2002;62:1256–1260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

