Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470
- PMID: 19660503
- PMCID: PMC2759695
- DOI: 10.1016/j.molbiopara.2009.07.008
Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470
Abstract
Microsporidia are protists that have been reported to cause infections in both vertebrates and invertebrates. They have emerged as human pathogens particularly in patients that are immunosuppressed and cases of gastrointestinal infection, encephalitis, keratitis, sinusitis, myositis and disseminated infection are well described in the literature. While benzimidazoles are active against many species of microsporidia, these drugs do not have significant activity against Enterocytozoon bieneusi. Fumagillin and its analogues have been demonstrated to have activity invitro and in animal models of microsporidiosis and human infections due to E. bieneusi. Fumagillin and its analogues inhibit methionine aminopeptidase type 2. Encephalitozoon cuniculi MetAP2 (EcMetAP2) was cloned and expressed as an active enzyme using a baculovirus system. The crystal structure of EcMetAP2 was determined with and without the bound inhibitors fumagillin and TNP-470. This structure classifies EcMetAP2 as a member of the MetAP2c family. The EcMetAP2 structure was used to generate a homology model of the E. bieneusi MetAP2. Comparison of microsporidian MetAP2 structures with human MetAP2 provides insights into the design of inhibitors that might exhibit specificity for microsporidian MetAP2.
Figures

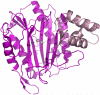

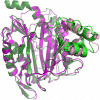
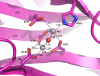
Similar articles
-
Investigations into microsporidian methionine aminopeptidase type 2: a therapeutic target for microsporidiosis.Folia Parasitol (Praha). 2005 May;52(1-2):182-92. doi: 10.14411/fp.2005.023. Folia Parasitol (Praha). 2005. PMID: 16004378 Free PMC article.
-
System for expression of microsporidian methionine amino peptidase type 2 (MetAP2) in the yeast Saccharomyces cerevisiae.Antimicrob Agents Chemother. 2006 Oct;50(10):3389-95. doi: 10.1128/AAC.00726-06. Epub 2006 Aug 17. Antimicrob Agents Chemother. 2006. PMID: 16917013 Free PMC article.
-
Homology modeling and calculation of the cobalt cluster charges of the Encephazlitozoon cuniculi methionine aminopeptidase, a potential target for drug design.Biophys Chem. 2003 Aug 1;105(1):29-43. doi: 10.1016/s0301-4622(03)00056-5. Biophys Chem. 2003. PMID: 12932577
-
Methionine aminopeptidases: Potential therapeutic target for microsporidia and other microbes.J Eukaryot Microbiol. 2024 Sep-Oct;71(5):e13036. doi: 10.1111/jeu.13036. Epub 2024 Jul 22. J Eukaryot Microbiol. 2024. PMID: 39036929 Review.
-
Therapeutic targets for the treatment of microsporidiosis in humans.Expert Opin Ther Targets. 2018 Nov;22(11):903-915. doi: 10.1080/14728222.2018.1538360. Epub 2018 Nov 1. Expert Opin Ther Targets. 2018. PMID: 30336698 Free PMC article. Review.
Cited by
-
Metallo-aminopeptidase inhibitors.Biochimie. 2010 Nov;92(11):1509-29. doi: 10.1016/j.biochi.2010.04.026. Epub 2010 May 10. Biochimie. 2010. PMID: 20457213 Free PMC article. Review.
-
Porphyrins inactivate Nosema spp. microsporidia.Sci Rep. 2018 Apr 3;8(1):5523. doi: 10.1038/s41598-018-23678-8. Sci Rep. 2018. PMID: 29615690 Free PMC article.
-
Discovery of novel antigiardiasis drug candidates.Antimicrob Agents Chemother. 2014 Dec;58(12):7303-11. doi: 10.1128/AAC.03834-14. Epub 2014 Sep 29. Antimicrob Agents Chemother. 2014. PMID: 25267663 Free PMC article.
-
Establishment and application of a loop-mediated isothermal amplification method based on MetAP2 gene for the detection of Nosema bombycis in silkworms (Bombyx mori).Front Vet Sci. 2025 Mar 10;12:1549224. doi: 10.3389/fvets.2025.1549224. eCollection 2025. Front Vet Sci. 2025. PMID: 40129576 Free PMC article.
-
The state of research for AIDS-associated opportunistic infections and the importance of sustaining smaller research communities.Eukaryot Cell. 2012 Feb;11(2):90-7. doi: 10.1128/EC.05143-11. Epub 2011 Dec 9. Eukaryot Cell. 2012. PMID: 22158712 Free PMC article. No abstract available.
References
-
- Sprague V, Becnel JJ, Hazard EI. Taxonomy of phylum microspora. Crit Rev Microbiol. 1992;18:285–395. - PubMed
-
- Wittner M, W. LM. The Microsporidia and Microsporidiosis. Vol. 577. ASM Press; Washington, D.C.: 1999.
-
- Takvorian PM, Cali A. Enzyme histochemical identification of the Golgi apparatus in the microsporidian, Glugea stephani. J Eukaryot Microbiol. 1994;41:63S–64S. - PubMed
-
- Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–9. - PubMed
-
- Keeling PJ. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet Biol. 2003;38:298–309. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

