BayesCall: A model-based base-calling algorithm for high-throughput short-read sequencing
- PMID: 19661376
- PMCID: PMC2765266
- DOI: 10.1101/gr.095299.109
BayesCall: A model-based base-calling algorithm for high-throughput short-read sequencing
Abstract
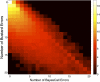
Extracting sequence information from raw images of fluorescence is the foundation underlying several high-throughput sequencing platforms. Some of the main challenges associated with this technology include reducing the error rate, assigning accurate base-specific quality scores, and reducing the cost of sequencing by increasing the throughput per run. To demonstrate how computational advancement can help to meet these challenges, a novel model-based base-calling algorithm, BayesCall, is introduced for the Illumina sequencing platform. Being founded on the tools of statistical learning, BayesCall is flexible enough to incorporate various features of the sequencing process. In particular, it can easily incorporate time-dependent parameters and model residual effects. This new approach significantly improves the accuracy over Illumina's base-caller Bustard, particularly in the later cycles of a sequencing run. For 76-cycle data on a standard viral sample, phiX174, BayesCall improves Bustard's average per-base error rate by approximately 51%. The probability of observing each base can be readily computed in BayesCall, and this probability can be transformed into a useful base-specific quality score with a high discrimination ability. A detailed study of BayesCall's performance is presented here.
Figures







References
-
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. - PubMed
-
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. - PubMed
-
- Li L, Speed T. An estimate of the crosstalk matrix in four-dye fluorescence-based DNA sequencing. Electrophoresis. 1999;20:1433–1442. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
