Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence
- PMID: 19662163
- PMCID: PMC2714078
- DOI: 10.1371/journal.pgen.1000592
Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence
Abstract
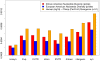
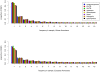
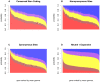
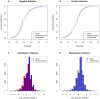
Analysis of polymorphism and divergence in the non-coding portion of the human genome yields crucial information about factors driving the evolution of gene regulation. Candidate cis-regulatory regions spanning more than 15,000 genes in 15 African Americans and 20 European Americans were re-sequenced and aligned to the chimpanzee genome in order to identify potentially functional polymorphism and to characterize and quantify departures from neutral evolution. Distortions of the site frequency spectra suggest a general pattern of selective constraint on conserved non-coding sites in the flanking regions of genes (CNCs). Moreover, there is an excess of fixed differences that cannot be explained by a Gamma model of deleterious fitness effects, suggesting the presence of positive selection on CNCs. Extensions of the McDonald-Kreitman test identified candidate cis-regulatory regions with high probabilities of positive and negative selection near many known human genes, the biological characteristics of which exhibit genome-wide trends that differ from patterns observed in protein-coding regions. Notably, there is a higher probability of positive selection in candidate cis-regulatory regions near genes expressed in the fetal brain, suggesting that a larger portion of adaptive regulatory changes has occurred in genes expressed during brain development. Overall we find that natural selection has played an important role in the evolution of candidate cis-regulatory regions throughout hominid evolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. - PubMed
-
- Taylor MS, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2:e30. doi: 10.1371/journal.pgen.0020030. - DOI - PMC - PubMed

