Operons
- PMID: 19662496
- PMCID: PMC2776167
- DOI: 10.1007/s00018-009-0114-3
Operons
Abstract
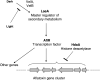
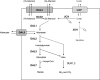
Operons (clusters of co-regulated genes with related functions) are common features of bacterial genomes. More recently, functional gene clustering has been reported in eukaryotes, from yeasts to filamentous fungi, plants, and animals. Gene clusters can consist of paralogous genes that have most likely arisen by gene duplication. However, there are now many examples of eukaryotic gene clusters that contain functionally related but non-homologous genes and that represent functional gene organizations with operon-like features (physical clustering and co-regulation). These include gene clusters for use of different carbon and nitrogen sources in yeasts, for production of antibiotics, toxins, and virulence determinants in filamentous fungi, for production of defense compounds in plants, and for innate and adaptive immunity in animals (the major histocompatibility locus). The aim of this article is to review features of functional gene clusters in prokaryotes and eukaryotes and the significance of clustering for effective function.
Figures








References
-
- Jacob F, Perrin D, Sanchez C, Monod J. L’operon: Groupe de genes a l’expression coordonne par un operateur. C R Acad Sci. 1960;245:1727–729.
-
- Jacob F, Monod J (1961) On the regulation of gene activity. In: Cold Spring Harbor Symposium Quantitative Biology 26, pp 193–211
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

