Complexity of the MSG gene family of Pneumocystis carinii
- PMID: 19664205
- PMCID: PMC2743713
- DOI: 10.1186/1471-2164-10-367
Complexity of the MSG gene family of Pneumocystis carinii
Abstract
Background: The relationship between the parasitic fungus Pneumocystis carinii and its host, the laboratory rat, presumably involves features that allow the fungus to circumvent attacks by the immune system. It is hypothesized that the major surface glycoprotein (MSG) gene family endows Pneumocystis with the capacity to vary its surface. This gene family is comprised of approximately 80 genes, which each are approximately 3 kb long. Expression of the MSG gene family is regulated by a cis-dependent mechanism that involves a unique telomeric site in the genome called the expression site. Only the MSG gene adjacent to the expression site is represented by messenger RNA. Several P. carinii MSG genes have been sequenced, which showed that genes in the family can encode distinct isoforms of MSG. The vast majority of family members have not been characterized at the sequence level.
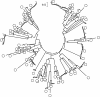
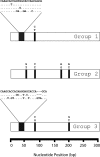
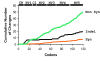
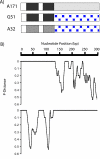
Results: The first 300 basepairs of MSG genes were subjected to analysis herein. Analysis of 581 MSG sequence reads from P. carinii genomic DNA yielded 281 different sequences. However, many of the sequence reads differed from others at only one site, a degree of variation consistent with that expected to be caused by error. Accounting for error reduced the number of truly distinct sequences observed to 158, roughly twice the number expected if the gene family contains 80 members. The size of the gene family was verified by PCR. The excess of distinct sequences appeared to be due to allelic variation. Discounting alleles, there were 73 different MSG genes observed. The 73 genes differed by 19% on average. Variable regions were rich in nucleotide differences that changed the encoded protein. The genes shared three regions in which at least 16 consecutive basepairs were invariant. There were numerous cases where two different genes were identical within a region that was variable among family members as a whole, suggesting recombination among family members.
Conclusion: A set of sequences that represents most if not all of the members of the P. carinii MSG gene family was obtained. The protein-changing nature of the variation among these sequences suggests that the family has been shaped by selection for protein variation, which is consistent with the hypothesis that the MSG gene family functions to enhance phenotypic variation among the members of a population of P. carinii.
Figures
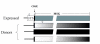

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

