The adult boar testicular and epididymal transcriptomes
- PMID: 19664223
- PMCID: PMC2738690
- DOI: 10.1186/1471-2164-10-369
The adult boar testicular and epididymal transcriptomes
Abstract
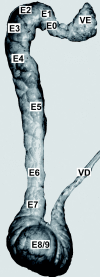
Background: Mammalians gamete production takes place in the testis but when they exit this organ, although spermatozoa have acquired a specialized and distinct morphology, they are immotile and infertile. It is only after their travel in the epididymis that sperm gain their motility and fertility. Epididymis is a crescent shaped organ adjacent to the testis that can be divided in three gross morphological regions, head (caput), body (corpus) and tail (cauda). It contains a long and unique convoluted tubule connected to the testis via the efferent ducts and finished by joining the vas deferens in its caudal part.
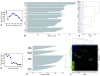
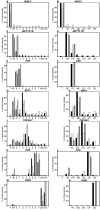
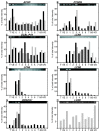
Results: In this study, the testis, the efferent ducts (vas efferens, VE), nine distinct successive epididymal segments and the deferent duct (vas deferens, VD) of four adult boars of known fertility were isolated and their mRNA extracted. The gene expression of each of these samples was analyzed using a pig generic 9 K nylon microarray (AGENAE program; GEO accession number: GPL3729) spotted with 8931 clones derived from normalized cDNA banks from different pig tissues including testis and epididymis. Differentially expressed transcripts were obtained with moderated t-tests and F-tests and two data clustering algorithms based either on partitioning around medoid (top down PAM) or hierarchical clustering (bottom up HCL) were combined for class discovery and gene expression analysis. Tissue clustering defined seven transcriptomic units: testis, vas efferens and five epididymal transcriptomic units. Meanwhile transcripts formed only four clusters related to the tissues. We have then used a specific statistical method to sort out genes specifically over-expressed (markers) in testis, VE or in each of the five transcriptomic units of the epididymis (including VD). The specific regional expression of some of these genes was further validated by PCR and Q-PCR. We also searched for specific pathways and functions using available gene ontology information.
Conclusion: This study described for the first time the complete transcriptomes of the testis, the epididymis, the vas efferens and the vas deferens on the same species. It described new genes or genes not yet reported over-expressed in these boar tissues, as well as new control mechanisms. It emphasizes and fulfilled the gap between studies done in rodents and human, and provides tools that will be useful for further studies on the biochemical processes responsible for the formation and maintain of the epididymal regionalization and the development of a fertile spermatozoa.
Figures






Similar articles
-
Impact of male fertility status on the transcriptome of the bovine epididymis.Mol Hum Reprod. 2017 Jun 1;23(6):355-369. doi: 10.1093/molehr/gax019. Mol Hum Reprod. 2017. PMID: 28379507
-
Profiling of relaxin and its receptor proteins in boar reproductive tissues and spermatozoa.Reprod Biol Endocrinol. 2015 May 20;13:46. doi: 10.1186/s12958-015-0043-y. Reprod Biol Endocrinol. 2015. PMID: 25990010 Free PMC article.
-
The epididymal transcriptome and proteome provide some insights into new epididymal regulations.J Androl. 2011 Nov-Dec;32(6):651-64. doi: 10.2164/jandrol.111.013086. Epub 2011 Jul 15. J Androl. 2011. PMID: 21764898 Review.
-
G protein-coupled estrogen receptor (GPER) in adult boar testes, epididymis and spermatozoa during epididymal maturation.Int J Biol Macromol. 2018 Sep;116:113-119. doi: 10.1016/j.ijbiomac.2018.05.015. Epub 2018 May 4. Int J Biol Macromol. 2018. PMID: 29730010
-
Development and use of surgical procedures to bypass selected regions of the mammalian epididymis: effects on sperm maturation.J Reprod Fertil Suppl. 1998;53:183-95. J Reprod Fertil Suppl. 1998. PMID: 10645277 Review.
Cited by
-
Network Analyses Predict Small RNAs That Might Modulate Gene Expression in the Testis and Epididymis of Bos indicus Bulls.Front Genet. 2021 Apr 30;12:610116. doi: 10.3389/fgene.2021.610116. eCollection 2021. Front Genet. 2021. PMID: 33995471 Free PMC article.
-
Characteristics of the Epididymal Luminal Environment Responsible for Sperm Maturation and Storage.Front Endocrinol (Lausanne). 2018 Feb 28;9:59. doi: 10.3389/fendo.2018.00059. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 29541061 Free PMC article. Review.
-
Sperm-Coating Beta-Defensin 126 Is a Dissociation-Resistant Dimer Produced by Epididymal Epithelium in the Bovine Reproductive Tract.Biol Reprod. 2016 Dec;95(6):121. doi: 10.1095/biolreprod.116.138719. Epub 2016 Oct 5. Biol Reprod. 2016. PMID: 27707712 Free PMC article.
-
Sperm Proteome after Interaction with Reproductive Fluids in Porcine: From the Ejaculation to the Fertilization Site.Int J Mol Sci. 2020 Aug 22;21(17):6060. doi: 10.3390/ijms21176060. Int J Mol Sci. 2020. PMID: 32842715 Free PMC article.
-
Identification and Functional Assignment of Genes Implicated in Sperm Maturation of Tibetan Sheep.Animals (Basel). 2023 May 6;13(9):1553. doi: 10.3390/ani13091553. Animals (Basel). 2023. PMID: 37174590 Free PMC article.
References
-
- Dacheux JL, Paquignon M. Relations between the fertilizing ability, motility and metabolism of epididymal spermatozoa. Reproduction, nutrition, development. 1980;20:1085–1099. - PubMed
-
- Robaire B, Hinton BT, Orgebin-Crist MC. The epididymis. In: JN K, editor. Physiology of reproduction. New York: Elsevier; 2006. pp. 1071–1148.
-
- Turner TT. Resorption versus secretion in the rat epididymis. J Reprod Fertil. 1984;72:509–514. - PubMed
-
- Turner TT. De Graaf's Thread: The Human Epididymis. J Androl. 2008;29:237–250. - PubMed
-
- Dacheux JL, Castella S, Gatti JL, Dacheux F. Epididymal cell secretory activities and the role of proteins in boar sperm maturation. Theriogenology. 2005;63:319–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

