Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes
- PMID: 19664233
- PMCID: PMC2743667
- DOI: 10.1186/1471-2148-9-194
Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes
Abstract
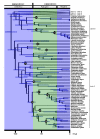
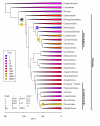
Background: One of the main explanations for the stunning diversity of teleost fishes (approximately 29,000 species, nearly half of all vertebrates) is that a fish-specific whole-genome duplication event (FSGD) in the ancestor to teleosts triggered their subsequent radiation. However, one critical assumption of this hypothesis, that diversification rates in teleosts increased soon after the acquisition of a duplicated genome, has never been tested.
Results: Here we show that one of three major diversification rate shifts within ray-finned fishes occurred at the base of the teleost radiation, as predicted by the FSGD hypothesis. We also find evidence for two rate increases that are much younger than the inferred age of the FSGD: one in the common ancestor of most ostariophysan fishes, and a second one in the common ancestor of percomorphs. The biodiversity contained within these two clades accounts for more than 88% of living fish species.
Conclusion: Teleosts diversified explosively in their early history and this burst of diversification may have been caused by genome duplication. However, the FSGD itself may be responsible for a little over 10% of living teleost biodiversity. ~88% of species diversity is derived from two relatively recent radiations of freshwater and marine fishes where genome duplication is not suspected. Genome duplications are a common event on the tree of life and have been implicated in the diversification of major clades like flowering plants, vertebrates, and gnathostomes. However our results suggest that the causes of diversification in large clades are likely to be complex and not easily ascribed to a single event, even a dramatic one such as a whole genome duplication.
Figures


References
-
- FISHBASE. World Wide Web electronic publication http://www.fishbase.org

