A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease
- PMID: 19664744
- PMCID: PMC2725265
- DOI: 10.1016/j.ajhg.2009.07.012
A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease
Abstract
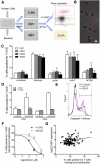
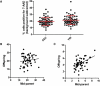
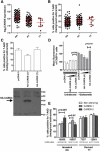
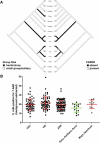
Recent progress in cataloguing common genetic variation has made possible genome-wide studies that are beginning to elucidate the causes and consequences of our genetic differences. Approaches that provide a mechanistic understanding of how genetic variants function to alter disease susceptibility and why they were substrates of natural selection would complement other approaches to human-genome analysis. Here we use a novel cell-based screen of bacterial infection to identify human variation in Salmonella-induced cell death. A loss-of-function allele of CARD8, a reported inhibitor of the proinflammatory protease caspase-1, was associated with increased cell death in vitro (p = 0.013). The validity of this association was demonstrated through overexpression of alternative alleles and RNA interference in cells of varying genotype. Comparison of mammalian CARD8 orthologs and examination of variation among different human populations suggest that the increase in infectious-disease burden associated with larger animal groups (i.e., herds and colonies), and possibly human population expansion, may have naturally selected for loss of CARD8. We also find that the loss-of-function CARD8 allele shows a modest association with an increased risk of systemic inflammatory response syndrome in a small study (p = 0.05). Therefore, a by-product of the selected benefit of loss of CARD8 could be increased inflammatory diseases. These results demonstrate the utility of genome-wide cell-based association screens with microbes in the identification of naturally selected variants that can impact human health.
Figures

References
-
- Edwards A.O., Ritter R., Abel K.J., Manning A., Panhuysen C., Farrer L.A. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. - PubMed
-
- Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. - PubMed
-
- Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. - PubMed
-
- Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

