Pyriform spidroin 1, a novel member of the silk gene family that anchors dragline silk fibers in attachment discs of the black widow spider, Latrodectus hesperus
- PMID: 19666476
- PMCID: PMC2781455
- DOI: 10.1074/jbc.M109.021378
Pyriform spidroin 1, a novel member of the silk gene family that anchors dragline silk fibers in attachment discs of the black widow spider, Latrodectus hesperus
Abstract
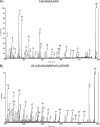
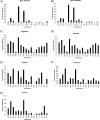
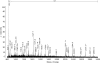
Spiders spin high performance threads that have diverse mechanical properties for specific biological applications. To better understand the molecular mechanism by which spiders anchor their threads to a solid support, we solubilized the attachment discs from black widow spiders and performed in-solution tryptic digests followed by MS/MS analysis to identify novel peptides derived from glue silks. Combining matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry and cDNA library screening, we isolated a novel member of the silk gene family called pysp1 and demonstrate that its protein product is assembled into the attachment disc silks. Alignment of the PySp1 amino acid sequence to other fibroins revealed conservation in the non-repetitive C-terminal region of the silk family. MS/MS analysis also confirmed the presence of MaSp1 and MaSp2, two important components of dragline silks, anchored within the attachment disc materials. Characterization of the ultrastructure of attachment discs using scanning electron microscopy studies support the localization of PySp1 to small diameter fibers embedded in a glue-like cement, which network with large diameter dragline silk threads, producing a strong, adhesive material. Consistent with elevated PySp1 mRNA levels detected in the pyriform gland, MS analysis of the luminal contents extracted from the pyriform gland after tryptic digestion support the assertion that PySp1 represents one of the major constituents manufactured in the pyriform gland. Taken together, our data demonstrate that PySp1 is spun into attachment disc silks to help affix dragline fibers to substrates, a critical function during spider web construction for prey capture and locomotion.
Figures



Similar articles
-
Egg case protein-1. A new class of silk proteins with fibroin-like properties from the spider Latrodectus hesperus.J Biol Chem. 2005 Jun 3;280(22):21220-30. doi: 10.1074/jbc.M412316200. Epub 2005 Mar 29. J Biol Chem. 2005. PMID: 15797873
-
Aciniform spidroin, a constituent of egg case sacs and wrapping silk fibers from the black widow spider Latrodectus hesperus.J Biol Chem. 2007 Nov 30;282(48):35088-97. doi: 10.1074/jbc.M705791200. Epub 2007 Oct 5. J Biol Chem. 2007. PMID: 17921147
-
Spider minor ampullate silk proteins are constituents of prey wrapping silk in the cob weaver Latrodectus hesperus.Biochemistry. 2008 Apr 22;47(16):4692-700. doi: 10.1021/bi800140q. Epub 2008 Apr 1. Biochemistry. 2008. PMID: 18376847
-
Molecular mechanisms of spider silk.Cell Mol Life Sci. 2006 Sep;63(17):1986-99. doi: 10.1007/s00018-006-6090-y. Cell Mol Life Sci. 2006. PMID: 16819558 Free PMC article. Review.
-
Spider silk: understanding the structure-function relationship of a natural fiber.Prog Mol Biol Transl Sci. 2011;103:131-85. doi: 10.1016/B978-0-12-415906-8.00007-8. Prog Mol Biol Transl Sci. 2011. PMID: 21999996 Review.
Cited by
-
Chromosome mapping of dragline silk genes in the genomes of widow spiders (Araneae, Theridiidae).PLoS One. 2010 Sep 21;5(9):e12804. doi: 10.1371/journal.pone.0012804. PLoS One. 2010. PMID: 20877726 Free PMC article.
-
A recombinant chimeric spider pyriform-aciniform silk with highly tunable mechanical performance.Mater Today Bio. 2024 Apr 27;26:101073. doi: 10.1016/j.mtbio.2024.101073. eCollection 2024 Jun. Mater Today Bio. 2024. PMID: 38711935 Free PMC article.
-
Numerical implementation of multiple peeling theory and its application to spider web anchorages.Interface Focus. 2015 Feb 6;5(1):20140051. doi: 10.1098/rsfs.2014.0051. Interface Focus. 2015. PMID: 25657835 Free PMC article.
-
Experimental strategies for the identification and characterization of adhesive proteins in animals: a review.Interface Focus. 2015 Feb 6;5(1):20140064. doi: 10.1098/rsfs.2014.0064. Interface Focus. 2015. PMID: 25657842 Free PMC article. Review.
-
Hybrid Spider Silk with Inorganic Nanomaterials.Nanomaterials (Basel). 2020 Sep 16;10(9):1853. doi: 10.3390/nano10091853. Nanomaterials (Basel). 2020. PMID: 32947954 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

