The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis
- PMID: 19666488
- PMCID: PMC2728962
- DOI: 10.1073/pnas.0906509106
The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis
Abstract
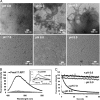
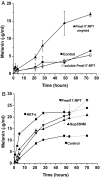
Pmel17 is a melanocyte protein necessary for eumelanin deposition 1 in mammals and found in melanosomes in a filamentous form. The luminal part of human Pmel17 includes a region (RPT) with 10 copies of a partial repeat sequence, pt.e.gttp.qv., known to be essential in vivo for filament formation. We show that this RPT region readily forms amyloid in vitro, but only under the mildly acidic conditions typical of the lysosome-like melanosome lumen, and the filaments quickly become soluble at neutral pH. Under the same mildly acidic conditions, the Pmel filaments promote eumelanin formation. Electron diffraction, circular dichroism, and solid-state NMR studies of Pmel17 filaments show that the structure is rich in beta sheet. We suggest that RPT is the amyloid core domain of the Pmel17 filaments so critical for melanin formation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sunde M, Kwan AH, Templeton MD, Beever RE, MacKay JP. Structural analysis of hydrophobins. Micron. 2008;39:773–784. - PubMed
-
- Podrabsky JE, Carpenter JF, Hand SC. Survival of water stress in annual fish embryos: Dehydration avoidance and egg amyloid fibers. Am J Physiol Regulatory Integrative Comp Physiol. 2001;280:R123–R131. - PubMed
-
- Iconomidou VA, Vriend G, Hamodrakas SJ. Amyloids protect the sildmoth oocyte and embryo. FEBS Lett. 2000;479:141–145. - PubMed
-
- Benkemoun L, Saupe SJ. Prion proteins as genetic material in fungi. Fungal Genet Biol. 2006;43:789–803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

