Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system
- PMID: 19666509
- PMCID: PMC2718047
- DOI: 10.1073/pnas.0901566106
Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system
Abstract
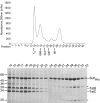
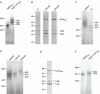
The Tat system transports folded proteins across the bacterial cytoplasmic membrane and the thylakoid membrane of plant chloroplasts. In Escherichia coli substrate proteins initially bind to the integral membrane TatBC complex which then recruits the protein TatA to effect translocation. Overproduction of TatBC and the substrate protein SufI in the absence of TatA led to the accumulation of TatBC-SufI complexes that could be purified using an affinity tag on the substrate. Three-dimensional structures of the TatBC-SufI complexes and unliganded TatBC were obtained by single-particle electron microscopy and random conical tilt reconstruction. Comparison of the structures shows that substrate molecules bind on the periphery of the TatBC complex and that substrate binding causes a significant reduction in diameter of the TatBC part of the complex. Although the TatBC complex contains multiple copies of the signal peptide-binding TatC protomer, purified TatBC-SufI complexes contain only 1 or 2 SufI molecules. Where 2 substrates are present in the TatBC-SufI complex, they are bound at adjacent sites. These observations imply that only certain TatC protomers within the complex interact with substrate or that there is a negative cooperativity of substrate binding. Similar TatBC-substrate complexes can be generated by an alternative in vitro reconstitution method and using a different substrate protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expressed at native levels.FEBS J. 2006 Dec;273(24):5656-68. doi: 10.1111/j.1742-4658.2006.05554.x. FEBS J. 2006. PMID: 17212781
-
The early mature part of bacterial twin-arginine translocation (Tat) precursor proteins contributes to TatBC receptor binding.J Biol Chem. 2018 May 11;293(19):7281-7299. doi: 10.1074/jbc.RA118.002576. Epub 2018 Mar 28. J Biol Chem. 2018. PMID: 29593092 Free PMC article.
-
The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex.J Mol Biol. 2005 Feb 11;346(1):295-305. doi: 10.1016/j.jmb.2004.11.047. Epub 2004 Dec 13. J Mol Biol. 2005. PMID: 15663945
-
Twin-arginine-dependent translocation of folded proteins.Philos Trans R Soc Lond B Biol Sci. 2012 Apr 19;367(1592):1029-46. doi: 10.1098/rstb.2011.0202. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22411976 Free PMC article. Review.
-
Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria.Biochim Biophys Acta. 2011 Mar;1808(3):876-84. doi: 10.1016/j.bbamem.2010.11.023. Epub 2010 Nov 29. Biochim Biophys Acta. 2011. PMID: 21126506 Review.
Cited by
-
Precursor-Receptor Interactions in the Twin Arginine Protein Transport Pathway Probed with a New Receptor Complex Preparation.Biochemistry. 2018 Mar 13;57(10):1663-1671. doi: 10.1021/acs.biochem.8b00026. Epub 2018 Feb 26. Biochemistry. 2018. PMID: 29460615 Free PMC article.
-
Signal Peptide Hydrophobicity Modulates Interaction with the Twin-Arginine Translocase.mBio. 2017 Aug 1;8(4):e00909-17. doi: 10.1128/mBio.00909-17. mBio. 2017. PMID: 28765221 Free PMC article.
-
New insights into the Tat protein transport cycle from characterizing the assembled Tat translocon.Mol Microbiol. 2022 Dec;118(6):637-651. doi: 10.1111/mmi.14984. Epub 2022 Oct 5. Mol Microbiol. 2022. PMID: 36151601 Free PMC article.
-
Routing of thylakoid lumen proteins by the chloroplast twin arginine transport pathway.Photosynth Res. 2018 Dec;138(3):289-301. doi: 10.1007/s11120-018-0567-z. Epub 2018 Aug 12. Photosynth Res. 2018. PMID: 30101370 Review.
-
Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells.PLoS One. 2013 Aug 2;8(8):e69488. doi: 10.1371/journal.pone.0069488. Print 2013. PLoS One. 2013. PMID: 23936332 Free PMC article.
References
-
- Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol. 2003;47:187–254. - PubMed
-
- Cline K, Theg SM. The Sec and Tat protein translocation pathways in chloroplasts. In: Dalbey RE, Koehler C, Tamanoi F, editors. The Enzymes, Molecular Machines Involved in Protein Transport across Cellular Membranes. Vol. XXV. San Diego, CA: Elsevier; 2007. pp. 455–485.
-
- Natale P, Brüser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim Biophys Acta. 2008;778:1735–1756. - PubMed
-
- Berks BC, Palmer T, Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol. 2005;8:174–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- D011140/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- C516144/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 072681/Z/03/Z/WT_/Wellcome Trust/United Kingdom
- BB/C516179/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D011140/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C516144/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G117/519/MRC_/Medical Research Council/United Kingdom
- BB/D011140/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D004578/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 079605/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

