Cell-type-specific isolation of ribosome-associated mRNA from complex tissues
- PMID: 19666516
- PMCID: PMC2728999
- DOI: 10.1073/pnas.0907143106
Cell-type-specific isolation of ribosome-associated mRNA from complex tissues
Abstract
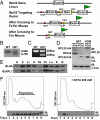
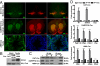
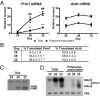
Gene profiling techniques allow the assay of transcripts from organs, tissues, and cells with an unprecedented level of coverage. However, most of these approaches are still limited by the fact that organs and tissues are composed of multiple cell types that are each unique in their patterns of gene expression. To identify the transcriptome from a single cell type in a complex tissue, investigators have relied upon physical methods to separate cell types or in situ hybridization and immunohistochemistry. Here, we describe a strategy to rapidly and efficiently isolate ribosome-associated mRNA transcripts from any cell type in vivo. We have created a mouse line, called RiboTag, which carries an Rpl22 allele with a floxed wild-type C-terminal exon followed by an identical C-terminal exon that has three copies of the hemagglutinin (HA) epitope inserted before the stop codon. When the RiboTag mouse is crossed to a cell-type-specific Cre recombinase-expressing mouse, Cre recombinase activates the expression of epitope-tagged ribosomal protein RPL22(HA), which is incorporated into actively translating polyribosomes. Immunoprecipitation of polysomes with a monoclonal antibody against HA yields ribosome-associated mRNA transcripts from specific cell types. We demonstrate the application of this technique in brain using neuron-specific Cre recombinase-expressing mice and in testis using a Sertoli cell Cre recombinase-expressing mouse.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
RiboTag: Ribosomal Tagging Strategy to Analyze Cell-Type-Specific mRNA Expression In Vivo.Curr Protoc Neurosci. 2019 Jun;88(1):e77. doi: 10.1002/cpns.77. Curr Protoc Neurosci. 2019. PMID: 31216392 Free PMC article.
-
Translatome Analyses Using Conditional Ribosomal Tagging in GABAergic Interneurons and Other Sparse Cell Types.Curr Protoc Neurosci. 2020 Jun;92(1):e93. doi: 10.1002/cpns.93. Curr Protoc Neurosci. 2020. PMID: 32584517 Free PMC article.
-
Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice.Genesis. 2002 Jul;33(3):114-8. doi: 10.1002/gene.10100. Genesis. 2002. PMID: 12124943
-
An Optimized Protocol for the Mapping of Cell Type-Specific Ribosome-Associated Transcript Isoforms from Small Mouse Brain Regions.Methods Mol Biol. 2022;2537:37-49. doi: 10.1007/978-1-0716-2521-7_3. Methods Mol Biol. 2022. PMID: 35895257
-
Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods.Methods Mol Biol. 2009;553:109-26. doi: 10.1007/978-1-60327-563-7_6. Methods Mol Biol. 2009. PMID: 19588103 Review.
Cited by
-
Activity-dependent translation dynamically alters the proteome of the perisynaptic astrocyte process.Cell Rep. 2022 Oct 18;41(3):111474. doi: 10.1016/j.celrep.2022.111474. Cell Rep. 2022. PMID: 36261025 Free PMC article.
-
Alterations in the intrinsic properties of striatal cholinergic interneurons after dopamine lesion and chronic L-DOPA.Elife. 2020 Jul 20;9:e56920. doi: 10.7554/eLife.56920. Elife. 2020. PMID: 32687053 Free PMC article.
-
Elongation inhibitors do not prevent the release of puromycylated nascent polypeptide chains from ribosomes.Elife. 2020 Aug 26;9:e60048. doi: 10.7554/eLife.60048. Elife. 2020. PMID: 32844746 Free PMC article.
-
Estrogen Regulation of the Molecular Phenotype and Active Translatome of AVPV Kisspeptin Neurons.Endocrinology. 2021 Sep 1;162(9):bqab080. doi: 10.1210/endocr/bqab080. Endocrinology. 2021. PMID: 33856454 Free PMC article.
-
Cell type-specific gene expression profiling in brain tissue: comparison between TRAP, LCM and RNA-seq.BMB Rep. 2015 Jul;48(7):388-94. doi: 10.5483/bmbrep.2015.48.7.218. BMB Rep. 2015. PMID: 25603796 Free PMC article.
References
-
- Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. - PubMed
-
- Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protoc. 2007;2:2924–2929. - PubMed
-
- Lobo MK, et al. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

