Structural organization of box C/D RNA-guided RNA methyltransferase
- PMID: 19666563
- PMCID: PMC2728976
- DOI: 10.1073/pnas.0905128106
Structural organization of box C/D RNA-guided RNA methyltransferase
Abstract
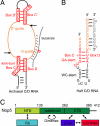
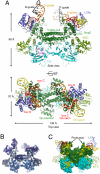
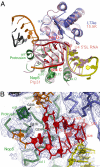
Box C/D guide RNAs are abundant noncoding RNAs that primarily function to direct the 2'-O-methylation of specific nucleotides by base-pairing with substrate RNAs. In archaea, a bipartite C/D RNA assembles with L7Ae, Nop5, and the methyltransferase fibrillarin into a modification enzyme with unique substrate specificity. Here, we determined the crystal structure of an archaeal C/D RNA-protein complex (RNP) composed of all 3 core proteins and an engineered half-guide RNA at 4 A resolution, as well as 2 protein substructures at higher resolution. The RNP structure reveals that the C-terminal domains of Nop5 in the dimeric complex provide symmetric anchoring sites for 2 L7Ae-associated kink-turn motifs of the C/D RNA. A prominent protrusion in Nop5 seems to be important for guide RNA organization and function and for discriminating the structurally related U4 snRNA. Multiple conformations of the N-terminal domain of Nop5 and its associated fibrillarin in different structures indicate the inherent flexibility of the catalytic module, suggesting that a swinging motion of the catalytic module is part of the enzyme mechanism. We also built a model of a native C/D RNP with substrate and fibrillarin in an active conformation. Our results provide insight into the overall organization and mechanism of action of C/D RNA-guided RNA methyltransferases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

