Negative membrane curvature as a cue for subcellular localization of a bacterial protein
- PMID: 19666580
- PMCID: PMC2726380
- DOI: 10.1073/pnas.0906851106
Negative membrane curvature as a cue for subcellular localization of a bacterial protein
Abstract
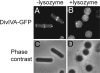
Bacterial proteins often localize to distinct sites within the cell, but the primary cues that dictate localization are largely unknown. Recent evidence has shown that positive membrane curvature can serve as a cue for localization of a peripheral membrane protein. Here we report that localization of the peripheral membrane protein DivIVA is determined in whole or in part by recognition of negative membrane curvature and that regions of the protein near the N and C terminus are important for localization. DivIVA, which is a cell division protein in Bacillus subtilis, localizes principally as a ring at nascent septa and secondarily to the less negatively curved, inside surface of the hemispherical poles of the cell. When cytokinesis is prevented, DivIVA redistributes itself to, and becomes markedly enriched at, the poles. When the rod-shaped cells are converted into spheres (protoplasts) by treatment with lysozyme, DivIVA adopts a largely uniform distribution around the cell. Recognition of membrane curvature by peripheral membrane proteins could be a general strategy for protein localization in bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. - PubMed
-
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–241. - PubMed
-
- Kim H, et al. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006;59:487–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

