Trypanosomatid genomes contain several subfamilies of ingi-related retroposons
- PMID: 19666780
- PMCID: PMC2756862
- DOI: 10.1128/EC.00183-09
Trypanosomatid genomes contain several subfamilies of ingi-related retroposons
Abstract
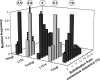
Retroposons are ubiquitous transposable elements found in the genomes of most eukaryotes, including trypanosomatids. The African and American trypanosomes (Trypanosoma brucei and Trypanosoma cruzi) contain long autonomous retroposons of the ingi clade (Tbingi and L1Tc, respectively) and short nonautonomous truncated versions (TbRIME and NARTc, respectively), as well as degenerate ingi-related retroposons devoid of coding capacity (DIREs). In contrast, Leishmania major contains only remnants of extinct retroposons (LmDIREs) and of short nonautonomous heterogeneous elements (LmSIDERs). We extend this comparative and evolutionary analysis of retroposons to the genomes of two other African trypanosomes (Trypanosoma congolense and Trypanosoma vivax) and another Leishmania sp. (Leishmania braziliensis). Three new potentially functional retroposons of the ingi clade have been identified: Tvingi in T. vivax and Tcoingi and L1Tco in T. congolense. T. congolense is the first trypanosomatid containing two classes of potentially active retroposons of the ingi clade. We analyzed sequences located upstream of these new long autonomous ingi-related elements, which code for the recognition site of the retroposon-encoded endonuclease. The closely related Tcoingi and Tvingi elements show the same conserved pattern, indicating that the Tcoingi- and Tvingi-encoded endonucleases share site specificity. Similarly, the conserved pattern previously identified upstream of L1Tc has also been detected at the same relative position upstream of L1Tco elements. A phylogenetic analysis of all ingi-related retroposons identified so far, including DIREs, clearly shows that several distinct subfamilies have emerged and coexisted, though in the course of trypanosomatid evolution, only a few have been maintained as active elements in modern trypanosomatid (sub)species.
Figures


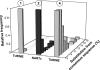
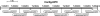
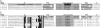



Similar articles
-
TSIDER1, a short and non-autonomous Salivarian trypanosome-specific retroposon related to the ingi6 subclade.Mol Biochem Parasitol. 2011 Sep;179(1):30-6. doi: 10.1016/j.molbiopara.2011.05.007. Epub 2011 Jun 1. Mol Biochem Parasitol. 2011. PMID: 21664383 Free PMC article.
-
Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements.Mol Biochem Parasitol. 2006 Feb;145(2):158-70. doi: 10.1016/j.molbiopara.2005.09.017. Epub 2005 Oct 10. Mol Biochem Parasitol. 2006. PMID: 16257065
-
Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome.BMC Genomics. 2009 May 22;10:240. doi: 10.1186/1471-2164-10-240. BMC Genomics. 2009. PMID: 19463167 Free PMC article.
-
Signalling the genome: the Ras-like small GTPase family of trypanosomatids.Trends Parasitol. 2005 Oct;21(10):447-50. doi: 10.1016/j.pt.2005.08.008. Trends Parasitol. 2005. PMID: 16112905 Review.
-
Repetitive elements in genomes of parasitic protozoa.Microbiol Mol Biol Rev. 2003 Sep;67(3):360-75, table of contents. doi: 10.1128/MMBR.67.3.360-375.2003. Microbiol Mol Biol Rev. 2003. PMID: 12966140 Free PMC article. Review.
Cited by
-
LINEs Contribute to the Origins of Middle Bodies of SINEs besides 3' Tails.Genome Biol Evol. 2018 Jan 1;10(1):370-379. doi: 10.1093/gbe/evy008. Genome Biol Evol. 2018. PMID: 29325122 Free PMC article.
-
Pr77 and L1TcRz: A dual system within the 5'-end of L1Tc retrotransposon, internal promoter and HDV-like ribozyme.Mob Genet Elements. 2012 Jan 1;2(1):1-7. doi: 10.4161/mge.19233. Mob Genet Elements. 2012. PMID: 22754746 Free PMC article.
-
The Trypanosomatid Pr77-hallmark contains a downstream core promoter element essential for transcription activity of the Trypanosoma cruzi L1Tc retrotransposon.BMC Genomics. 2016 Feb 9;17:105. doi: 10.1186/s12864-016-2427-6. BMC Genomics. 2016. PMID: 26861854 Free PMC article.
-
Repetitive elements in parasitic protozoa.BMC Biol. 2010 May 24;8:64. doi: 10.1186/1741-7007-8-64. BMC Biol. 2010. PMID: 20529240 Free PMC article.
-
TSIDER1, a short and non-autonomous Salivarian trypanosome-specific retroposon related to the ingi6 subclade.Mol Biochem Parasitol. 2011 Sep;179(1):30-6. doi: 10.1016/j.molbiopara.2011.05.007. Epub 2011 Jun 1. Mol Biochem Parasitol. 2011. PMID: 21664383 Free PMC article.
References
-
- Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, S. E. Melville, and N. M. El-Sayed. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. - PubMed
-
- Bohne, A., F. Brunet, D. Galiana-Arnoux, C. Schultheis, and J. N. Volff. 2008. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 16:203-215. - PubMed
-
- Boucher, N., Y. Wu, C. Dumas, M. Dube, D. Sereno, M. Breton, and B. Papadopoulou. 2002. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 277:19511-19520. - PubMed
-
- Bringaud, F., D. C. Bartholomeu, G. Blandin, A. Delcher, T. Baltz, N. M. El-Sayed, and E. Ghedin. 2006. The Trypanosoma cruzi L1Tc and NARTc non-LTR retrotransposons show relative site-specificity for insertion. Mol. Biol. Evol. 23:411-420. - PubMed
-
- Bringaud, F., N. Biteau, E. Zuiderwijk, M. Berriman, N. M. El-Sayed, E. Ghedin, S. E. Melville, N. Hall, and T. Baltz. 2004. The ingi and RIME non-LTR retrotransposons are not randomly distributed in the genome of Trypanosoma brucei. Mol. Biol. Evol. 21:520-528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

