Coupled RNA processing and transcription of intergenic primary microRNAs
- PMID: 19667074
- PMCID: PMC2756881
- DOI: 10.1128/MCB.00664-09
Coupled RNA processing and transcription of intergenic primary microRNAs
Abstract
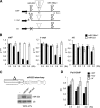
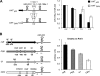
The first step in microRNA (miRNA) biogenesis occurs in the nucleus and is mediated by the Microprocessor complex containing the RNase III-like enzyme Drosha and its cofactor DGCR8. Here we show that the 5'-->3' exonuclease Xrn2 associates with independently transcribed miRNAs and, in combination with Drosha processing, attenuates transcription in downstream regions. We suggest that, after Drosha cleavage, a torpedo-like mechanism acts on nascent long precursor miRNAs, whereby Xrn2 exonuclease degrades the RNA polymerase II-associated transcripts inducing its release from the template. While involved in primary transcript termination, this attenuation effect does not restrict clustered miRNA expression, which, in the majority of cases, is separated by short spacers. We also show that transcripts originating from a miRNA promoter are retained on the chromatin template and are more efficiently processed than those produced from mRNA or snRNA Pol II-dependent promoters. These data imply that coupling between transcription and processing promotes efficient expression of independently transcribed miRNAs.
Figures
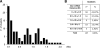



References
-
- Aguilera, A. 2005. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 17:242-250. - PubMed
-
- Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. - PubMed
-
- Buratowski, S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17:257-261. - PubMed
-
- Chakrabarti, S. K., J. C. James, and R. G. Mirmira. 2002. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J. Biol. Chem. 277:13286-13293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
