TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II
- PMID: 19667075
- PMCID: PMC2756882
- DOI: 10.1128/MCB.00637-09
TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II
Abstract
The function of human TFIIH-associated Cdk7 in RNA polymerase II (Pol II) transcription and C-terminal domain (CTD) phosphorylation was investigated in analogue-sensitive Cdk7(as/as) mutant cells where the kinase can be inhibited without disrupting TFIIH. We show that both Cdk7 and Cdk9/PTEFb contribute to phosphorylation of Pol II CTD Ser5 residues on transcribed genes. Cdk7 is also a major kinase of CTD Ser7 on Pol II at the c-fos and U snRNA genes. Furthermore, TFIIH and recombinant Cdk7-CycH-Mat1 as well as recombinant Cdk9-CycT1 phosphorylated CTD Ser7 and Ser5 residues in vitro. Inhibition of Cdk7 in vivo suppressed the amount of Pol II accumulated at 5' ends on several genes including c-myc, p21, and glyceraldehyde-3-phosphate dehydrogenase genes, indicating reduced promoter-proximal pausing or polymerase "leaking" into the gene. Consistent with a 5' pausing defect, Cdk7 inhibition reduced recruitment of the negative elongation factor NELF at start sites. A role of Cdk7 in regulating elongation is further suggested by enhanced histone H4 acetylation and diminished histone H4 trimethylation on lysine 36-two marks of elongation-within genes when the kinase was inhibited. Consistent with a new role for TFIIH at 3' ends, it was detected within genes and 3'-flanking regions, and Cdk7 inhibition delayed pausing and transcription termination.
Figures
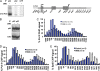
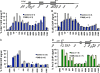

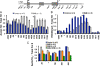

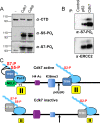
References
-
- Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. - PubMed
-
- Akoulitchev, S., T. P. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. - PubMed
-
- Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17:251-256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
