A role for polypyrimidine tract binding protein in the establishment of focal adhesions
- PMID: 19667078
- PMCID: PMC2756873
- DOI: 10.1128/MCB.00590-09
A role for polypyrimidine tract binding protein in the establishment of focal adhesions
Abstract
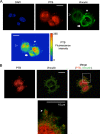
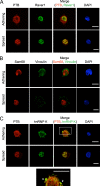
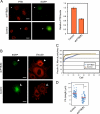
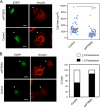
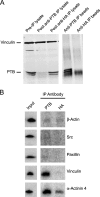
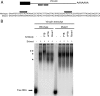
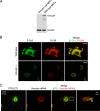
Polypyrimidine tract binding protein (PTB) is a widely expressed RNA binding protein. In the nucleus PTB regulates the splicing of alternative exons, while in the cytoplasm it can affect mRNA stability, translation, and localization. Here we demonstrate that PTB transiently localizes to the cytoplasm and to protrusions in the cellular edge of mouse embryo fibroblasts during adhesion to fibronectin and the early stages of cell spreading. This cytoplasmic PTB is associated with transcripts encoding the focal adhesion scaffolding proteins vinculin and alpha-actinin 4. We demonstrate that vinculin mRNA colocalizes with PTB to cytoplasmic protrusions and that PTB depletion reduces vinculin mRNA at the cellular edge and limits the size of focal adhesions. The loss of PTB also alters cell morphology and limits the ability of cells to spread after adhesion. These data indicate that during the initial stages of cell adhesion, PTB shuttles from the nucleus to the cytoplasm and influences focal adhesion formation through coordinated control of scaffolding protein mRNAs.
Figures


References
-
- Bomsztyk, K., O. Denisenko, and J. Ostrowski. 2004. hnRNP K: one protein multiple processes. Bioessays 26:629-638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
