Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism
- PMID: 19667173
- PMCID: PMC2731842
- DOI: 10.1073/pnas.0904489106
Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism
Abstract
We provide a demonstration in humans of the principle of pharmacometabonomics by showing a clear connection between an individual's metabolic phenotype, in the form of a predose urinary metabolite profile, and the metabolic fate of a standard dose of the widely used analgesic acetaminophen. Predose and postdose urinary metabolite profiles were determined by (1)H NMR spectroscopy. The predose spectra were statistically analyzed in relation to drug metabolite excretion to detect predose biomarkers of drug fate and a human-gut microbiome cometabolite predictor was identified. Thus, we found that individuals having high predose urinary levels of p-cresol sulfate had low postdose urinary ratios of acetaminophen sulfate to acetaminophen glucuronide. We conclude that, in individuals with high bacterially mediated p-cresol generation, competitive O-sulfonation of p-cresol reduces the effective systemic capacity to sulfonate acetaminophen. Given that acetaminophen is such a widely used and seemingly well-understood drug, this finding provides a clear demonstration of the immense potential and power of the pharmacometabonomic approach. However, we expect many other sulfonation reactions to be similarly affected by competition with p-cresol and our finding also has important implications for certain diseases as well as for the variable responses induced by many different drugs and xenobiotics. We propose that assessing the effects of microbiome activity should be an integral part of pharmaceutical development and of personalized health care. Furthermore, we envisage that gut bacterial populations might be deliberately manipulated to improve drug efficacy and to reduce adverse drug reactions.
Conflict of interest statement
Conflict of interest statement: T.A.C., J.C.L., J.R.E., and J.K.N. are inventors on a relevant patent application from which financial gain might be derived. T.A.C., J.C.L., and J.K.N. might also benefit financially from the future placement of related analytical or research contracts.
Figures
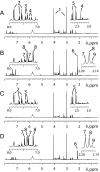
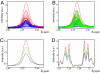

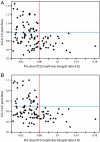
Comment in
-
Drugs, bugs, and personalized medicine: pharmacometabonomics enters the ring.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14187-8. doi: 10.1073/pnas.0907721106. Epub 2009 Aug 19. Proc Natl Acad Sci U S A. 2009. PMID: 19706501 Free PMC article. No abstract available.
References
-
- Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science. 1999;286:487–491. - PubMed
-
- Evans WE, Johnson JA. Pharmacogenomics: The inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9–39. - PubMed
-
- Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. - PubMed
-
- Pagliarulo V, Datar RH, Cote RJ. Role of genetic and expression profiling in pharmacogenomics: The changing face of patient management. Curr Issues Mol Biol. 2002;4:101–110. - PubMed
-
- Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

