Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance
- PMID: 19667176
- PMCID: PMC2731839
- DOI: 10.1073/pnas.0813319106
Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance
Abstract
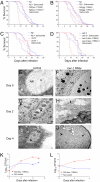
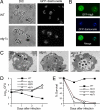
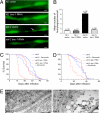
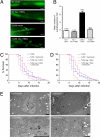
A conserved insulin-like pathway modulates both aging and pathogen resistance in Caenorhabditis elegans. However, the specific innate effector functions that mediate this pathogen resistance are largely unknown. Autophagy, a lysosomal degradation pathway, plays a role in controlling intracellular bacterial pathogen infections in cultured cells, but less is known about its role at the organismal level. We examined the effects of autophagy gene inactivation on Salmonella enterica Serovar Typhimurium (Salmonella typhimurium) infection in 2 model organisms, Caenorhabditis elegans and Dictyostelium discoideum. In both organisms, genetic inactivation of the autophagy pathway increases bacterial intracellular replication, decreases animal lifespan, and results in apoptotic-independent death. In C. elegans, genetic knockdown of autophagy genes abrogates pathogen resistance conferred by a loss-of-function mutation, daf-2(e1370), in the insulin-like tyrosine kinase receptor or by over-expression of the DAF-16 FOXO transcription factor. Thus, autophagy genes play an essential role in host defense in vivo against an intracellular bacterial pathogen and mediate pathogen resistance in long-lived mutant nematodes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Gavazzi G, Krause KH. Ageing and infection. Lancet Infect Dis. 2002;2:659–666. - PubMed
-
- Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. - PubMed
-
- Garsin D, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. - PubMed
-
- Melendez A, et al. Autophagy genes are essential for dauer development and lifespan extension in C. elegans. Science. 2003;301:1387–1391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

