Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells
- PMID: 19668214
- PMCID: PMC3987895
- DOI: 10.1038/ng.428
Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells
Abstract
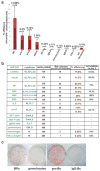
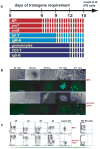
The reprogramming of somatic cells into induced pluripotent stem (iPS) cells upon overexpression of the transcription factors Oct4, Sox2, Klf4 and cMyc is inefficient. It has been assumed that the somatic differentiation state provides a barrier for efficient reprogramming; however, direct evidence for this notion is lacking. Here, we tested the potential of mouse hematopoietic cells at different stages of differentiation to be reprogrammed into iPS cells. We show that hematopoietic stem and progenitor cells give rise to iPS cells up to 300 times more efficiently than terminally differentiated B and T cells do, yielding reprogramming efficiencies of up to 28%. Our data provide evidence that the differentiation stage of the starting cell has a critical influence on the efficiency of reprogramming into iPS cells. Moreover, we identify hematopoietic progenitors as an attractive cell type for applications of iPS cell technology in research and therapy.
Figures




References
-
- Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. - PubMed
-
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. - PubMed
-
- Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. - PubMed
-
- Liao J, et al. Generation of Induced Pluripotent Stem Cell Lines from Adult Rat Cells. Cell Stem Cell. 2008 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

